6JJ3
 
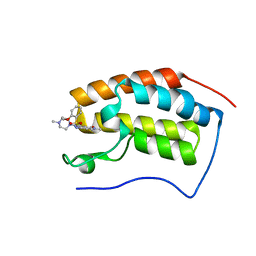 | | BRD4 in complex with 138A | | Descriptor: | 2-methoxy-N-[2-methyl-6-(4-methylpiperazin-1-yl)-3-oxidanylidene-2,7-diazatricyclo[6.3.1.0^{4,12}]dodeca-1(12),4,6,8,10-pentaen-9-yl]benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xu, J, Chen, Y, Jiang, F, Zhu, J. | | Deposit date: | 2019-02-25 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Discovery of Benzo[cd]indol-2(1H)-ones and Pyrrolo[4,3,2-de]quinolin-2(1H)-ones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with Selectivity for the First Bromodomain with Potential High Efficiency against Acute Gouty Arthritis.
J.Med.Chem., 62, 2019
|
|
3ZZT
 
 | |
6JBH
 
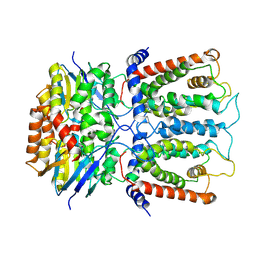 | | Cryo-EM structure and transport mechanism of a wall teichoic acid ABC transporter | | Descriptor: | TarG, TarH | | Authors: | Chen, L, Hou, W.T, Fan, T, Li, Y.H, Liu, B.H, Jiang, Y.L, Sun, L.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-electron Microscopy Structure and Transport Mechanism of a Wall Teichoic Acid ABC Transporter.
Mbio, 11, 2020
|
|
3ZZU
 
 | |
4ADN
 
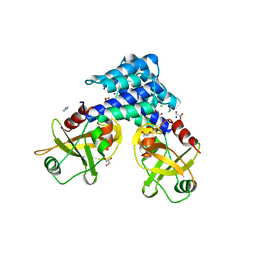 | | Fusidic acid resistance protein FusB | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FAR1, ... | | Authors: | Guo, X, Peisker, K, Backbro, K, Chen, Y, Kiran, R.K, Sanyal, S, Selmer, M. | | Deposit date: | 2011-12-31 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Function of Fusb: An Elongation Factor G-Binding Fusidic Acid Resistance Protein Active in Ribosomal Translocation and Recycling
Open Biol., 2, 2012
|
|
3UYI
 
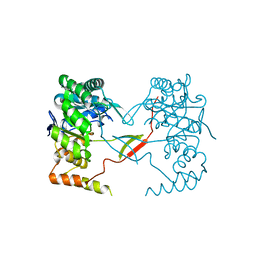 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | Perakine reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
3V0T
 
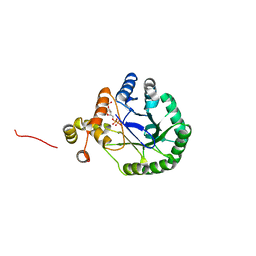 | | Crystal Structure of Perakine Reductase, Founder Member of a Novel AKR Subfamily with Unique Conformational Changes during NADPH Binding | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Perakine Reductase | | Authors: | Sun, L, Chen, Y, Rajendran, C, Panjikar, S, Mueller, U, Wang, M, Rosenthal, C, Mindnich, R, Penning, T.M, Stoeckigt, J. | | Deposit date: | 2011-12-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.333 Å) | | Cite: | Crystal structure of perakine reductase, founding member of a novel aldo-keto reductase (AKR) subfamily that undergoes unique conformational changes during NADPH binding.
J.Biol.Chem., 287, 2012
|
|
5F59
 
 | | The crystal structure of MLL3 SET domain | | Descriptor: | Histone-lysine N-methyltransferase 2C, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-04 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F6L
 
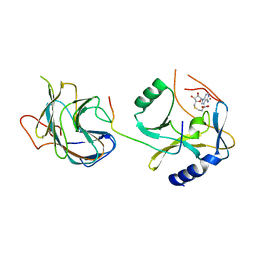 | | The crystal structure of MLL1 (N3861I/Q3867L) in complex with RbBP5 and Ash2L | | Descriptor: | Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
5F5E
 
 | |
5F6K
 
 | | Crystal structure of the MLL3-Ash2L-RbBP5 complex | | Descriptor: | Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Li, Y, Lei, M, Chen, Y. | | Deposit date: | 2015-12-06 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Structural basis for activity regulation of MLL family methyltransferases.
Nature, 530, 2016
|
|
4RDK
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis b tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
4RDL
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
4QSG
 
 | | Crystal structure of gas vesicle protein GvpF from Microcystis aeruginosa | | Descriptor: | Gas vesicle protein | | Authors: | Xu, B.Y, Dai, Y.N, Zhou, K, Ren, Y.M, Liu, Y.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-07-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the gas vesicle protein GvpF from the cyanobacterium Microcystis aeruginosa.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4RDJ
 
 | | Crystal structure of Norovirus Boxer P domain | | Descriptor: | Capsid | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
4PQG
 
 | | Crystal structure of the pneumococcal O-GlcNAc transferase GtfA in complex with UDP and GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosyltransferase Gtf1, URIDINE-5'-DIPHOSPHATE | | Authors: | Shi, W.W, Jiang, Y.L, Zhu, F, Yang, Y.H, Wu, H, Ren, Y.M, Chen, Y, Zhou, C.Z. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Novel O-Linked N-Acetyl-d-glucosamine (O-GlcNAc) Transferase, GtfA, Reveals Insights into the Glycosylation of Pneumococcal Serine-rich Repeat Adhesins.
J.Biol.Chem., 289, 2014
|
|
6JPE
 
 | | Crystal structure of FGFR4 kinase domain with irreversible inhibitor 1 | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[6-[2-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]amino]pyridin-3-yl]pyrimidin-4-yl]amino]-3-methyl-phenyl]prop-2-enamide, SULFATE ION | | Authors: | Chen, X, Dai, S, Zhou, Z, Chen, Y. | | Deposit date: | 2019-03-26 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Development of a Potent and Specific FGFR4 Inhibitor for the Treatment of Hepatocellular Carcinoma.
J.Med.Chem., 63, 2020
|
|
4QLA
 
 | | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori | | Descriptor: | Juvenile hormone epoxide hydrolase, PENTAETHYLENE GLYCOL | | Authors: | Zhou, K, Jia, N, Hu, C, Jiang, Y.L, Yang, J.P, Chen, Y, Li, S, Zhou, C.Z. | | Deposit date: | 2014-06-11 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of juvenile hormone epoxide hydrolase from the silkworm Bombyx mori.
Proteins, 82, 2014
|
|
2QIK
 
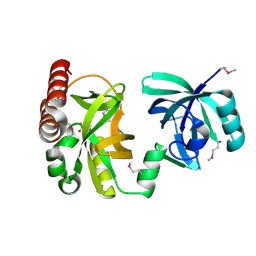 | | Crystal structure of YkqA from Bacillus subtilis. Northeast Structural Genomics Target SR631 | | Descriptor: | CITRIC ACID, UPF0131 protein ykqA | | Authors: | Benach, J, Chen, Y, Forouhar, F, Seetharaman, J, Baran, M.C, Cunningham, K, Ma, L.-C, Owens, L, Chen, C.X, Rong, X, Janjua, H, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of YkqA from Bacillus subtilis.
To be Published
|
|
4GDX
 
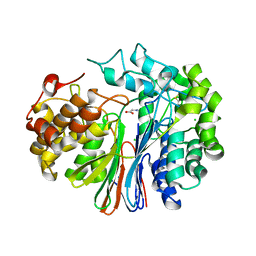 | | Crystal Structure of Human Gamma-Glutamyl Transpeptidase--Glutamate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
5F2E
 
 | | Crystal Structure of small molecule ARS-853 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-[4-[2-[[4-chloranyl-5-(1-methylcyclopropyl)-2-oxidanyl-phenyl]amino]ethanoyl]piperazin-1-yl]azetidin-1-yl]prop-2-en-1-one, GLYCEROL, GLYCINE, ... | | Authors: | Patricelli, M.P, Janes, M.R, Li, L.-S, Hansen, R, Peters, U, Kessler, L.V, Chen, Y, Kucharski, J.M, Feng, J, Ely, T, Chen, J.H, Firdaus, S.J, Babbar, A, Ren, P, Liu, Y. | | Deposit date: | 2015-12-01 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State.
Cancer Discov, 6, 2016
|
|
4IIK
 
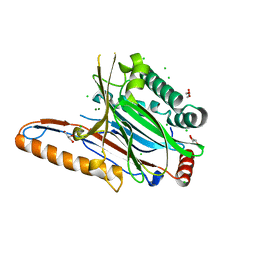 | | Legionella pneumophila effector | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tascon, I, Chen, Y, Neunuebel, M.R, Rojas, A.L, Machner, M.P, Hierro, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for Rab1 De-AMPylation by the Legionella pneumophila Effector SidD
Plos Pathog., 9, 2013
|
|
4IIP
 
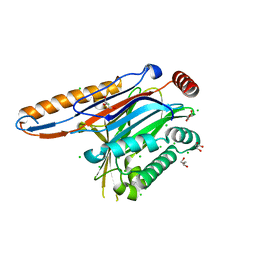 | | Legionella pneumophila effector | | Descriptor: | Adenosine monophosphate-protein hydrolase SidD, CHLORIDE ION, GLYCEROL | | Authors: | Tascon, I, Chen, Y, Neunuebel, M.R, Rojas, A.L, Machner, M.P, Hierro, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-06-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Rab1 De-AMPylation by the Legionella pneumophila Effector SidD
Plos Pathog., 9, 2013
|
|
4FJ4
 
 | | Crystal structure of the protein Q9HRE7 complexed with mercury from Halobacterium salinarium at the resolution 2.1A, Northeast Structural Genomics Consortium target HsR50 | | Descriptor: | ETHYL MERCURY ION, SODIUM ION, Uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Vorobiev, S.M, Seetharaman, J, Janjua, J, Xiao, R, Cunningham, K, Maglaqui, M, Owens, L.A, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-11 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Northeast Structural Genomics Consortium Target HsR50
To be Published
|
|
4GG2
 
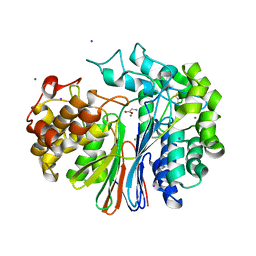 | | The crystal structure of glutamate-bound human gamma-glutamyltranspeptidase 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | West, M.B, Chen, Y, Wickham, S, Heroux, A, Cahill, K, Hanigan, M.H, Mooers, B.H.M. | | Deposit date: | 2012-08-04 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Novel Insights into Eukaryotic gamma-Glutamyltranspeptidase 1 from the Crystal Structure of the Glutamate-bound Human Enzyme.
J.Biol.Chem., 288, 2013
|
|
