4GQH
 
 | | The Conformations and Interactions of the Four-Layer Aggregate Revealed by X-ray Crystallography Diffraction Implied the Importance of Peptides at Opposite Ends in Their Assemblies | | Descriptor: | Capsid protein | | Authors: | Li, X.Y, Song, B.A, Hu, D.Y, Chen, X, Wang, Z.C, Zeng, M.J, Yu, D.D, Chen, Z, Jin, L.H, Yang, S. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The Conformations and Interactions of the Four-Layer Aggregate Revealed by X-ray Crystallography Diffraction Implied the Importance of Peptides at Opposite Ends in Their Assemblies
To be Published
|
|
4D8P
 
 | | Structural and functional studies of the trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) molecule | | Descriptor: | GLYCEROL, HLA-DQA1 protein, Peptide from Gamma-gliadin,HLA class II histocompatibility antigen, ... | | Authors: | Kim, C.-Y, Hotta, K, Mathews, I.I, Chen, X. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural and functional studies of trans-encoded HLA-DQ2.3 (DQA1*03:01/DQB1*02:01) protein molecule
J.Biol.Chem., 287, 2012
|
|
1QES
 
 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*GP*UP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
1QET
 
 | | TANDEM GU MISMATCHES IN RNA, NMR, 30 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*AP*UP*GP*UP*CP*C)-3') | | Authors: | Mcdowell, J.A, He, L, Chen, X, Turner, D.H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-06-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of the structural basis for thermodynamic stabilities of tandem GU wobble pairs: NMR structures of (rGGAGUUCC)2 and (rGGAUGUCC)2.
Biochemistry, 36, 1997
|
|
1R1V
 
 | | Crystal structure of the metal-sensing transcriptional repressor CzrA from Staphylococcus aureus in the Zn2-form | | Descriptor: | ZINC ION, repressor protein | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R22
 
 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB (C14S/C61S/C121S mutant) in the Zn2alpha5-form | | Descriptor: | Transcriptional repressor smtB, ZINC ION | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R1U
 
 | | Crystal structure of the metal-sensing transcriptional repressor CzrA from Staphylococcus aureus in the apo-form | | Descriptor: | repressor protein | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R23
 
 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB in the Zn1-form (one Zn(II) per dimer) | | Descriptor: | Transcriptional repressor smtB, ZINC ION | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
1R1T
 
 | | Crystal structure of the cyanobacterial metallothionein repressor SmtB in the apo-form | | Descriptor: | Transcriptional repressor smtB | | Authors: | Eicken, C, Pennella, M.A, Chen, X, Koshlap, K.M, VanZile, M.L, Sacchettini, J.C, Giedroc, D.P. | | Deposit date: | 2003-09-25 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A metal-ligand-mediated intersubunit allosteric switch in related SmtB/ArsR zinc sensor proteins.
J.Mol.Biol., 333, 2003
|
|
7RR5
 
 | | Structure of ribosomal complex bound with Rbg1/Tma46 | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Zeng, F, Li, X, Pires-Alves, M, Chen, X, Hawk, C.W, Jin, H. | | Deposit date: | 2021-08-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conserved heterodimeric GTPase Rbg1/Tma46 promotes efficient translation in eukaryotic cells.
Cell Rep, 37, 2021
|
|
7RSL
 
 | | Seipin forms a flexible cage at lipid droplet formation sites | | Descriptor: | Seipin | | Authors: | Arlt, H, Sui, X, Folger, B, Adams, C, Chen, X, Remme, R, Hamprecht, F.A, DiMaio, F, Liao, M, Goodman, J.M, Farese Jr, R.V, Walther, T.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-02-09 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Seipin forms a flexible cage at lipid droplet formation sites.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1S8K
 
 | | Solution Structure of BmKK4, A Novel Potassium Channel Blocker from Scorpion Buthus martensii Karsch, 25 structures | | Descriptor: | Toxin BmKK4 | | Authors: | Zhang, N, Chen, X, Li, M, Cao, C, Wang, Y, Hu, G, Wu, H. | | Deposit date: | 2004-02-02 | | Release date: | 2005-02-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK4, the first member of subfamily alpha-KTx 17 of scorpion toxins
Biochemistry, 43, 2004
|
|
1SVL
 
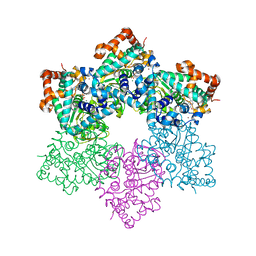 | | Co-crystal structure of SV40 large T antigen helicase domain and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
6VDK
 
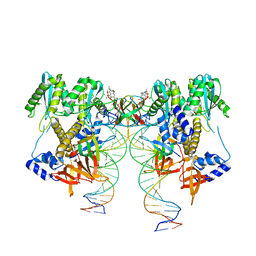 | | CryoEM structure of HIV-1 conserved Intasome Core | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Li, M, Chen, X, Craigie, R. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Peptide Derived from Lens Epithelium-Derived Growth Factor Stimulates HIV-1 DNA Integration and Facilitates Intasome Structural Studies.
J.Mol.Biol., 432, 2020
|
|
5KOC
 
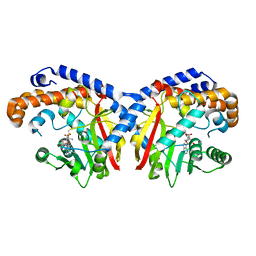 | | Pavine N-methyltransferase in complex with S-adenosylmethionine pH 7 | | Descriptor: | Pavine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Torres, M.A, Hoffarth, E, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KOK
 
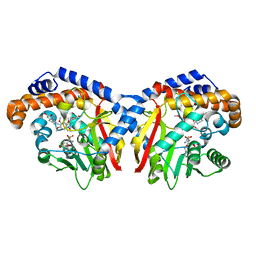 | | Pavine N-methyltransferase in complex with Tetrahydropapaverine and S-adenosylhomocysteine pH 7.25 | | Descriptor: | (1~{R})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, (1~{S})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, Pavine N-methyltransferase, ... | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-30 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
3VCB
 
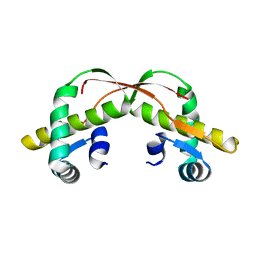 | | C425S mutant of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
5KPC
 
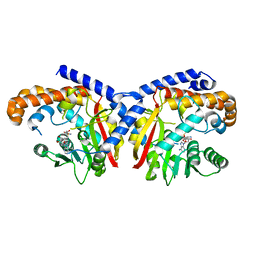 | | Pavine N-methyltransferase H206A mutant in complex with S-adenosylmethionine pH 6 | | Descriptor: | Pavine N-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-07-03 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KN4
 
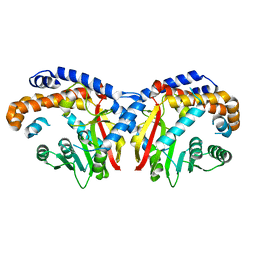 | | Pavine N-methyltransferase apoenzyme pH 6.0 | | Descriptor: | Pavine N-methyltransferase | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-06-27 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
5KPG
 
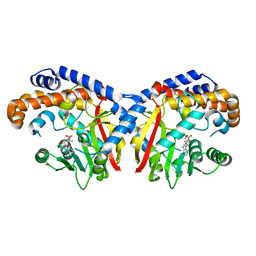 | | Pavine N-methyltransferase in complex with S-adenosylhomocysteine pH 7 | | Descriptor: | Pavine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Torres, M.A, Hoffarth, E, Eugenio, L, Savtchouk, J, Chen, X, Morris, J, Facchini, P.J, Ng, K.K.S. | | Deposit date: | 2016-07-04 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Studies of Pavine N-Methyltransferase from Thalictrum flavum Reveal Novel Insights into Substrate Recognition and Catalytic Mechanism.
J.Biol.Chem., 291, 2016
|
|
1SVM
 
 | | Co-crystal structure of SV40 large T antigen helicase domain and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
1SVO
 
 | | Structure of SV40 large T antigen helicase domain | | Descriptor: | ZINC ION, large T antigen | | Authors: | Gai, D, Zhao, R, Finkielstein, C.V, Chen, X.S. | | Deposit date: | 2004-03-29 | | Release date: | 2004-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen.
Cell(Cambridge,Mass.), 119, 2004
|
|
3VC8
 
 | | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59 | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Xu, X, Lou, Z, Ma, Y, Chen, X, Yang, Z, Tong, X, Zhao, Q, Xu, Y, Deng, H, Bartlam, M, Rao, Z. | | Deposit date: | 2012-01-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the C-terminal cytoplasmic domain of non-structural protein 4 from mouse hepatitis virus A59.
Plos One, 4, 2009
|
|
8XTR
 
 | | Comamonas testosteroni KF-1 circularly permuted group II intron 2S state | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, RNA (219-MER), ... | | Authors: | Wang, L, Xie, J.H, Zhang, C, Zou, J, Huang, Z, Shang, S, Chen, X, Yang, Y, Liu, J, Dong, H, Huang, D, Su, Z. | | Deposit date: | 2024-01-11 | | Release date: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis of circularly permuted group II intron self-splicing.
Nat.Struct.Mol.Biol., 2025
|
|
6P3Y
 
 | | Crystal Structure of Full Length APOBEC3G E/Q (pH 7.4) | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ZINC ION | | Authors: | Yang, H.J, Li, S.X, Chen, X.S. | | Deposit date: | 2019-05-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Understanding the structural basis of HIV-1 restriction by the full length double-domain APOBEC3G.
Nat Commun, 11, 2020
|
|
