4QDK
 
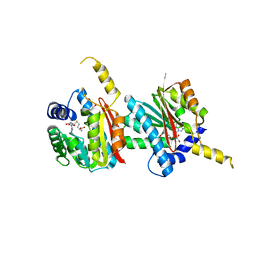 | |
4ZD3
 
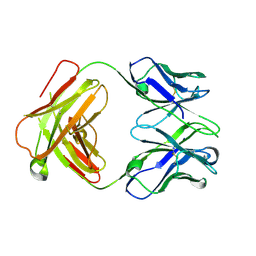 | | Structure of a transglutaminase 2-specific autoantibody Fab fragment | | Descriptor: | 679-14-14E06 Fab fragment heavy chain, 679-14-14E06 Fab fragment light chain | | Authors: | Chen, X, Dalhus, B, Hnida, K, Iversen, R, Sollid, L.M. | | Deposit date: | 2015-04-16 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Antigen Recognition by Transglutaminase 2-specific Autoantibodies in Celiac Disease.
J.Biol.Chem., 290, 2015
|
|
7K1J
 
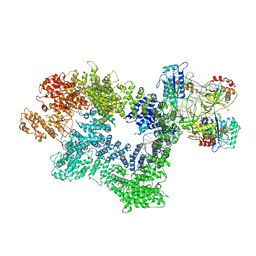 | | CryoEM structure of inactivated-form DNA-PK (Complex III) | | Descriptor: | DNA (5'-D(*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-07 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
7K1N
 
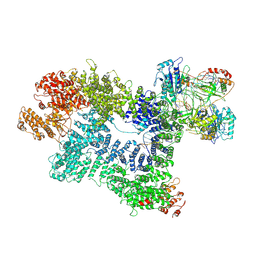 | | CryoEM structure of inactivated-form DNA-PK (Complex V) | | Descriptor: | DNA (5'-D(P*AP*AP*GP*CP*AP*GP*TP*AP*GP*AP*GP*CP*A)-3'), DNA (5'-D(P*GP*CP*AP*TP*GP*CP*TP*CP*TP*AP*CP*TP*GP*CP*TP*TP*CP*GP*AP*TP*AP*TP*CP*G)-3'), DNA-dependent protein kinase catalytic subunit, ... | | Authors: | Chen, X, Gellert, M, Yang, W. | | Deposit date: | 2020-09-08 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of an activated DNA-PK and its implications for NHEJ.
Mol.Cell, 81, 2021
|
|
5W1X
 
 | | Crystal Structure of Humanpapillomavirus18 (HPV18) Capsid L1 Pentamers Bound to Heparin Oligosaccharides | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, ... | | Authors: | Chen, X.S, Dasgupta, J. | | Deposit date: | 2017-06-05 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.374 Å) | | Cite: | Structural basis of oligosaccharide receptor recognition by human papillomavirus.
J. Biol. Chem., 286, 2011
|
|
3DR9
 
 | | Increased Distal Histidine Conformational Flexibility in the Deoxy Form of Dehaloperoxidase from Amphitrite ornata | | Descriptor: | Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Chen, X, de Serrano, V.S, Betts, L, Franzen, S. | | Deposit date: | 2008-07-10 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Distal histidine conformational flexibility in dehaloperoxidase from Amphitrite ornata.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3FUS
 
 | | Improved Structure of the Unliganded Simian Immunodeficiency Virus gp120 Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, X, Poon, B, Wang, Q, Ma, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
8QH2
 
 | | Crystal structure of chimeric UAP1L1 | | Descriptor: | UDP-N-acetylhexosamine pyrophosphorylase,UDP-N-acetylhexosamine pyrophosphorylase-like protein 1 | | Authors: | Chen, X, Yan, K, Bartual, S, van Aalten, D. | | Deposit date: | 2023-09-06 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | UAP1L1 is an active pyrophosphorylase with an active site disulfide regulating protein thermostability
To Be Published
|
|
4ZHJ
 
 | |
2NBV
 
 | |
2N3T
 
 | |
2N3W
 
 | |
474D
 
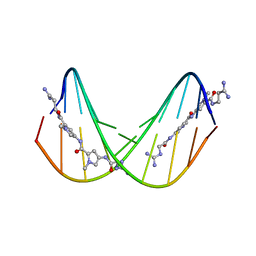 | | A NOVEL END-TO-END BINDING OF TWO NETROPSINS TO THE DNA DECAMER D(CCCCCIIIII)2 | | Descriptor: | DNA (5'-D(*CP*CP*CP*(CBR)P*CP*IP*IP*IP*IP*I)-3'), NETROPSIN | | Authors: | Chen, X, Rao, S.T, Sekar, K, Sundaralingam, M. | | Deposit date: | 1998-01-14 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel end-to-end binding of two netropsins to the DNA decamers d(CCCCCIIIII)2, d(CCCBr5CCIIIII)2and d(CBr5CCCCIIIII)2.
Nucleic Acids Res., 26, 1998
|
|
2NBW
 
 | | Solution structure of the Rpn1 T1 site with the Rad23 UBL domain | | Descriptor: | 26S proteasome regulatory subunit RPN1, UV excision repair protein RAD23 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2016-03-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome.
Structure, 24, 2016
|
|
2N3V
 
 | |
2NBU
 
 | |
2KQZ
 
 | |
2KR0
 
 | |
1CN3
 
 | |
2N3U
 
 | |
8KFO
 
 | | Crystal structure of BSA in complex with B3 | | Descriptor: | 6-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]-~{N},~{N}-dimethyl-naphthalen-2-amine, Albumin | | Authors: | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Bioinspired two-stage assembled photosensitive protein engineering for tumor-specific mitochondrial targeted phototherapy
To Be Published
|
|
6MUN
 
 | | Structure of hRpn10 bound to UBQLN2 UBL | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4, Ubiquilin-2 | | Authors: | Chen, X, Walters, K.J. | | Deposit date: | 2018-10-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome.
J.Mol.Biol., 431, 2019
|
|
6OER
 
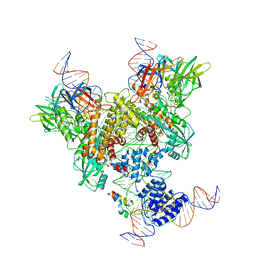 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA2) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8WQ3
 
 | | Crystal structure of the C-terminal RRM domain of an RBP | | Descriptor: | CHLORIDE ION, RNA-binding protein 45 | | Authors: | Chen, X, Jiang, M, Yang, Z, Chen, X, Wei, Q, Guo, S, Wang, M. | | Deposit date: | 2023-10-10 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural basis for RNA recognition by the C-terminal RRM domain of human RBM45.
J.Biol.Chem., 300, 2024
|
|
8WQ5
 
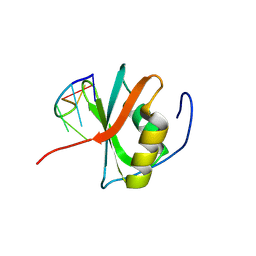 | | Crystal structure of the C-terminal RRM domain of an RBP in complex with ssDNA | | Descriptor: | DNA (5'-D(*GP*AP*CP*GP*CP*AP*G)-3'), GLYCEROL, RNA-binding protein 45 | | Authors: | Chen, X, Yang, Z, Chen, X, Wei, Q, Guo, S, Jiang, M, Wang, M. | | Deposit date: | 2023-10-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for RNA recognition by the C-terminal RRM domain of human RBM45.
J.Biol.Chem., 300, 2024
|
|
