4DNV
 
 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2013-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.764 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
2OD1
 
 | | Solution structure of the MYND domain from human AML1-ETO | | 分子名称: | Protein CBFA2T1, ZINC ION | | 著者 | Liu, Y.Z, Chen, W, Gaudet, J, Cheney, M.D, Roudaia, L, Cierpicki, T, Klet, R.C, Hartman, K, Laue, T.M, Speck, N.A, Bushweller, J.H. | | 登録日 | 2006-12-21 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity.
Cancer Cell, 11, 2007
|
|
2ODD
 
 | | Solution structure of the MYND domain from AML1-ETO complexed with SMRT, a corepressor | | 分子名称: | Protein CBFA2T1, SMRT, ZINC ION | | 著者 | Liu, Y.Z, Chen, W, Gaudet, J, Cheney, M.D, Roudaia, L, Cierpicki, T, Klet, R.C, Hartman, K, Laue, T.M, Speck, N.A, Bushweller, J.H. | | 登録日 | 2006-12-22 | | 公開日 | 2007-06-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for recognition of SMRT/N-CoR by the MYND domain and its contribution to AML1/ETO's activity.
Cancer Cell, 11, 2007
|
|
5Y2V
 
 | | Strcutrue of the full-length CcmR complexed with 2-OG from Synechocystis PCC6803 | | 分子名称: | 2-OXOGLUTARIC ACID, PHOSPHATE ION, Rubisco operon transcriptional regulator | | 著者 | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | 登録日 | 2017-07-27 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5Y2W
 
 | | Structure of Synechocystis PCC6803 CcmR regulatory domain in complex with 2-PG | | 分子名称: | 2-PHOSPHOGLYCOLIC ACID, Rubisco operon transcriptional regulator | | 著者 | Jiang, Y.L, Wang, X.P, Sun, H, Cheng, W, Cao, D.D, Han, S.J, Li, W.F, Chen, Y, Zhou, C.Z. | | 登録日 | 2017-07-27 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Coordinating carbon and nitrogen metabolic signaling through the cyanobacterial global repressor NdhR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6NHW
 
 | | Structure of the transmembrane domain of the Death Receptor 5 - Dimer of Trimer | | 分子名称: | Tumor necrosis factor receptor superfamily member 10B | | 著者 | Chou, J.J, Pan, L, Fu, Q, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H. | | 登録日 | 2018-12-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
6NHY
 
 | | Structure of the transmembrane domain of the Death Receptor 5 mutant (G217Y) - Trimer Only | | 分子名称: | Tumor necrosis factor receptor superfamily member 10B | | 著者 | Chou, J.J, Pan, L, Zhao, L, Chen, W, Piai, A, Fu, T, Wu, H, Liu, Z. | | 登録日 | 2018-12-24 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Higher-Order Clustering of the Transmembrane Anchor of DR5 Drives Signaling.
Cell, 176, 2019
|
|
4P5B
 
 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br dUMP | | 分子名称: | 5-BROMO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | 登録日 | 2014-03-15 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.274 Å) | | 主引用文献 | Crystal structure of a UMP/dUMP methylase PolB form Streptomyces cacaoi bound with 5-Br dUMP
To Be Published
|
|
4P5A
 
 | | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi bound with 5-Br UMP | | 分子名称: | 5-BROMO-URIDINE-5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, Thymidylate synthase ThyX | | 著者 | Li, Y, Chen, W, Li, J, Xia, Z, Deng, Z, Zhou, J. | | 登録日 | 2014-03-15 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of a UMP/dUMP methylase PolB from Streptomyces cacaoi with 5-Br UMP
To Be Published
|
|
4JD6
 
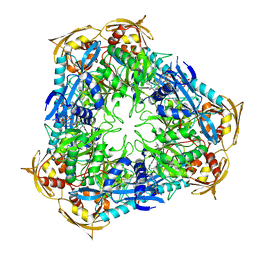 | | Crystal structure of Mycobacterium tuberculosis Eis in complex with coenzyme A and tobramycin | | 分子名称: | COENZYME A, Enhanced intracellular survival protein, TOBRAMYCIN | | 著者 | Biswas, T, Chen, W, Garneau-Tsodikova, S, Tsodikov, O.V. | | 登録日 | 2013-02-23 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Chemical and structural insights into the regioversatility of the aminoglycoside acetyltransferase eis.
Chembiochem, 14, 2013
|
|
5ZRZ
 
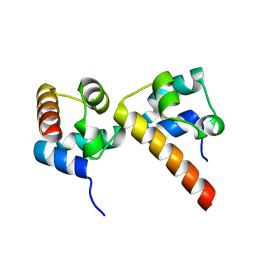 | | Crystal Structure of EphA5/SAMD5 Complex | | 分子名称: | Ephrin type-A receptor 5, Sterile alpha motif domain-containing protein 5 | | 著者 | Wang, Y, Shang, Y, Li, J, Chen, W, Li, G, Wan, J, Liu, W, Zhang, M. | | 登録日 | 2018-04-25 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Specific Eph receptor-cytoplasmic effector signaling mediated by SAM-SAM domain interactions.
Elife, 7, 2018
|
|
5IPI
 
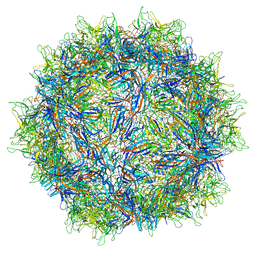 | | Structure of Adeno-associated virus type 2 VLP | | 分子名称: | Capsid protein VP1 | | 著者 | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | 登録日 | 2016-03-09 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
5IPK
 
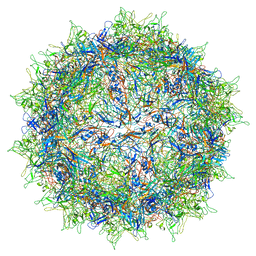 | | Structure of the R432A variant of Adeno-associated virus type 2 VLP | | 分子名称: | Capsid protein VP1 | | 著者 | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | 登録日 | 2016-03-09 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
6IZJ
 
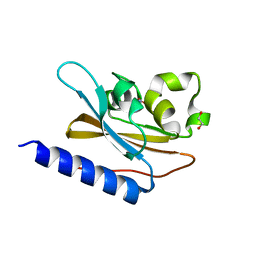 | | Structural characterization of mutated NreA protein in nitrate binding site from Staphylococcus aureus | | 分子名称: | 1,2-ETHANEDIOL, NITRATE ION, NreA | | 著者 | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | 登録日 | 2018-12-19 | | 公開日 | 2020-01-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
6K2H
 
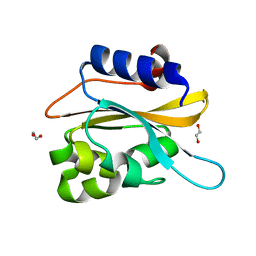 | | structural characterization of mutated NreA protein in nitrate binding site from staphylococcus aureus. | | 分子名称: | 1,2-ETHANEDIOL, NreA | | 著者 | Sangare, L, Chen, W, Wang, C, Chen, X, Wu, M, Zhang, X, Zang, J. | | 登録日 | 2019-05-14 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into the conformational change of Staphylococcus aureus NreA at C-terminus.
Biotechnol.Lett., 42, 2020
|
|
2M9J
 
 | | NMR solution structure of Pin1 WW domain mutant 6-1g | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | 登録日 | 2013-06-10 | | 公開日 | 2013-06-26 | | 最終更新日 | 2020-07-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9F
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1g | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | 登録日 | 2013-06-07 | | 公開日 | 2013-06-26 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9E
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | 登録日 | 2013-06-07 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9I
 
 | | NMR solution structure of Pin1 WW domain variant 6-1 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | 著者 | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | 登録日 | 2013-06-10 | | 公開日 | 2013-06-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2OIG
 
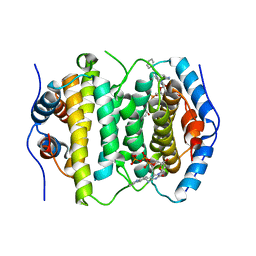 | | Crystal structure of RS21-C6 core segment and dm5CTP complex | | 分子名称: | 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), RS21-C6 | | 著者 | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | 登録日 | 2007-01-11 | | 公開日 | 2007-03-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
2OIE
 
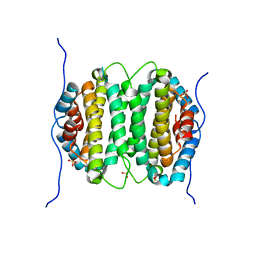 | | Crystal structure of RS21-C6 core segment RSCUT | | 分子名称: | RS21-C6, SULFATE ION | | 著者 | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | 登録日 | 2007-01-10 | | 公開日 | 2007-03-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
6M0W
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with the AGAA PAM | | 分子名称: | CRISPR-associated endonuclease Cas9 1, DNA (28-MER), DNA (5'-D(*AP*AP*AP*GP*AP*AP*GP*C)-3'), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-23 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.76 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
6M0X
 
 | | Crystal structure of Streptococcus thermophilus Cas9 in complex with AGGA PAM | | 分子名称: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-23 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.561 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
6M0V
 
 | | Crsytal structure of streptococcus thermophilus Cas9 in complex with the GGAA PAM | | 分子名称: | BARIUM ION, CRISPR-associated endonuclease Cas9 1, DNA (28-MER), ... | | 著者 | Zhang, Y, Zhang, H, Xu, X, Wang, Y, Chen, W, Wang, Y, Wu, Z, Tang, N, Wang, Y, Zhao, S, Gan, J, Ji, Q. | | 登録日 | 2020-02-22 | | 公開日 | 2020-09-09 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Catalytic-state structure and engineering of Streptococcus thermophilus Cas9
Nat Catal, 2020
|
|
