7E3X
 
 | | Crystal structure of SDR family NAD(P)-dependent oxidoreductase from exiguobacterium | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Chen, L, Tang, J, Yuan, S, Zhang, F, Chen, S. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-guided evolution of a ketoreductase forefficient and stereoselective bioreduction of bulkyalpha-aminobeta-keto esters
Catalysis Science And Technology, 2021
|
|
7EL8
 
 | |
7ENS
 
 | |
7ELT
 
 | |
7ENT
 
 | |
7DB7
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound GDI05-001 | | Descriptor: | 1-[3-[2-(1H-indol-3-yl)ethylsulfamoyl]phenyl]-3-(1,3-thiazol-2-yl)urea, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7EV2
 
 | |
7DAW
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase | | Descriptor: | Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, SULFATE ION | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7D47
 
 | | Crystal structure of SARS-CoV-2 Papain-like protease C111S | | Descriptor: | CALCIUM ION, Non-structural protein 3, ZINC ION | | Authors: | Wu, K.-P, Chen, S.-K, Lu, Y.-C, Huang, Y.-C.J, Lee, M.-H. | | Deposit date: | 2020-09-22 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of SARS-CoV-2 Papain-like protease
To Be Published
|
|
7DB8
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound PF-3845 | | Descriptor: | N-pyridin-3-yl-4-[[3-[5-(trifluoromethyl)pyridin-2-yl]oxyphenyl]methyl]piperidine-1-carboxamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7EV3
 
 | |
7DT1
 
 | | The structure of Lactobacillus fermentum 4,6-alpha-Glucanotransferase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W.K, Yong, Y.H, Wu, L, Chen, S, Zhou, J.H, Wu, J. | | Deposit date: | 2021-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43002272 Å) | | Cite: | Characterization of a new 4,6-alpha-glucanotransferase from Limosilactobacillus fermentum NCC 3057 with ability of synthesizing low molecular mass isomalto-/maltopolysaccharide
Food Biosci, 46, 2022
|
|
7DK2
 
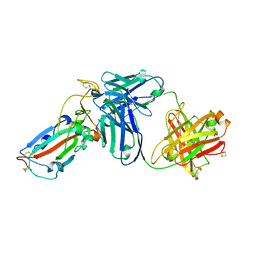 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW07 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MW07 heavy chain, MW07 light chain, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M, Chen, S. | | Deposit date: | 2020-11-22 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Architectural versatility of spike neutralization by a SARS-CoV-2 antibody
To Be Published
|
|
6IEW
 
 | |
5C3N
 
 | |
7OMT
 
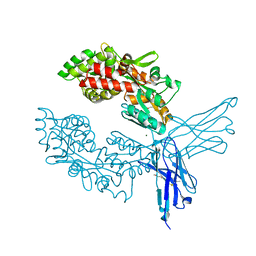 | | Crystal structure of ProMacrobody 21 with bound maltose | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7A8V
 
 | | Crystal structure of Polysaccharide monooxygenase from P.verruculosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Lytic polysaccharide monooxygenase, ... | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2020-08-31 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94961083 Å) | | Cite: | Crystal structure of Polysaccharide monooxygenase from P.verruculosum
To Be Published
|
|
7OMM
 
 | | Cryo-EM structure of N. gonorhoeae LptDE in complex with ProMacrobodies (MBPs have not been built de novo) | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ProMacrobody 21,Maltodextrin-binding protein, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
6AIJ
 
 | | Cyclodextrin glycosyltransferase from Paenibacillus macerans mutant N603D | | Descriptor: | CALCIUM ION, Cyclomaltodextrin glucanotransferase | | Authors: | Li, C.M, Ban, X.F, Li, Z.F, Li, Y.L, Cheng, S.D, Zhang, C.Y, Jin, T.C, Gu, Z.B. | | Deposit date: | 2018-08-24 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Cyclodextrin glycosyltransferase from Paenibacillus macerans mutant N603D
To Be Published
|
|
6YON
 
 | | Crystal structure of Endoglucanase S127C/A165C from Penicillium verruculosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase, SULFATE ION | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Endoglucanase S127C/A165C from Penicillium verruculosum
To Be Published
|
|
6T9F
 
 | | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24847341 Å) | | Cite: | CRYSTAL STRUCTURE OF EN ENDOGLUCANASE S308P FROM PENICILLIUM VERRUCULOSUM
To Be Published
|
|
6T9G
 
 | | CRYSTAL STRUCTURE OF AN ENDOGLUCANASE D213A FROM PENICILLIUM VERRUCULOSUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29620671 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENDOGLUCANASE D213A FROM PENICILLIUM VERRUCULOSUM
To Be Published
|
|
5I6S
 
 | | Crystal structure of an endoglucanase from Penicillium verruculosum | | Descriptor: | beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, endoglucanase | | Authors: | Nemashklaov, V, Vakhrusheva, A, Tishchenko, S, Gabdulkhakov, A, Kravchenko, O, Gusakov, A, Sinitsyn, A. | | Deposit date: | 2016-02-16 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an endoglucanase from Penicillium verruculosum
To Be Published
|
|
4NB7
 
 | | Crystal Structure of Two-Domain Laccase from Streptomyces LIvidans AC1709 in complex with azide after 180 min soaking | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, AZIDE ION, ... | | Authors: | Gabdulkhakov, A, Tischenko, S, Yurevich, L, Lisov, A, Leontievsky, A. | | Deposit date: | 2013-10-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Two-Domain Laccase from Streptomyces Lividans AC1709 in complex with azide after 180 min soaking
To be Published
|
|
4N8U
 
 | | Two-Domain Laccase from Streptomyces viridochromogenes at 2.4 A resolution AC629 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Gabdulkhakov, A, Tischenko, S, Yurevich, L, Lisov, A, Leontievsky, A. | | Deposit date: | 2013-10-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two-Domain Laccase from Streptomyces viridochromogenes at 2.4 A resolution AC629
To be Published
|
|
