4EP5
 
 | | Thermus thermophilus RuvC structure | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, GLYCEROL, SULFATE ION | | Authors: | Chen, L, Shi, K, Yin, Z.Q, Aihara, H. | | Deposit date: | 2012-04-17 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution.
Nucleic Acids Res., 41, 2013
|
|
8GZ3
 
 | |
4EAG
 
 | | Co-crystal structure of an chimeric AMPK core with ATP | | Descriptor: | 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EAI
 
 | | Co-crystal structure of an AMPK core with AMP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EAJ
 
 | | Co-crystal of AMPK core with AMP soaked with ATP | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Chen, L, Wang, J, Zhang, Y.-Y, Yan, S.F, Neumann, D, Schlattner, U, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2012-03-22 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | AMP-activated protein kinase undergoes nucleotide-dependent conformational changes
Nat.Struct.Mol.Biol., 19, 2012
|
|
4EP4
 
 | | Thermus thermophilus RuvC structure | | Descriptor: | Crossover junction endodeoxyribonuclease RuvC, GLYCEROL, MAGNESIUM ION | | Authors: | Chen, L, Shi, K, Yin, Z.Q, Aihara, H. | | Deposit date: | 2012-04-17 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural asymmetry in the Thermus thermophilus RuvC dimer suggests a basis for sequential strand cleavages during Holliday junction resolution.
Nucleic Acids Res., 41, 2013
|
|
3WQW
 
 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with a(GlcN)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Chen, L, Zhou, Y, Yang, Q. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fully deacetylated chitooligosaccharides act as efficient glycoside hydrolase family 18 chitinase inhibitors.
J.Biol.Chem., 289, 2014
|
|
3WQV
 
 | | Crystal structure of Ostrinia furnacalis Group I chitinase catalytic domain in complex with a(GlcN)5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Chen, L, Zhou, Y, Yang, Q. | | Deposit date: | 2014-02-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Fully deacetylated chitooligosaccharides act as efficient glycoside hydrolase family 18 chitinase inhibitors.
J.Biol.Chem., 289, 2014
|
|
2I0X
 
 | | Hypothetical protein PF1117 from Pyrococcus furiosus | | Descriptor: | Hypothetical protein PF1117 | | Authors: | Chen, L.Q, Fu, Z.-Q, Liu, Z.-J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-08-11 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Hypothetical Protein Pf1117 from Pyrococcus furiosus
To be Published
|
|
2HJD
 
 | |
5CCU
 
 | | Crystal structure of endoglycoceramidase I from Rhodococ-cus equi | | Descriptor: | 1,2-ETHANEDIOL, Putative secreted endoglycosylceramidase, SODIUM ION | | Authors: | Chen, L. | | Deposit date: | 2015-07-02 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Insights into the Broad Substrate Specificity of a Novel Endoglycoceramidase I Belonging to a New Subfamily of GH5 Glycosidases
J. Biol. Chem., 292, 2017
|
|
3IDX
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C222 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fab b13 heavy chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
3IDY
 
 | | Crystal structure of HIV-gp120 core in complex with CD4-binding site antibody b13, space group C2221 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab b13 heavy chain, Fab b13 light chain, ... | | Authors: | Chen, L, Kwon, Y.D, Zhou, T, Wu, X, O'Dell, S, Cavacini, L, Hessell, A.J, Pancera, M, Tang, M, Xu, L, Yang, Z.Y, Zhang, M.Y, Arthos, J, Burton, D.R, Dimitrov, D.S, Nabel, G.J, Posner, M, Sodroski, J, Wyatt, R, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2009-07-22 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120.
Science, 326, 2009
|
|
6BV3
 
 | | Crystal structure of porcine aminopeptidase-N with Leucine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV1
 
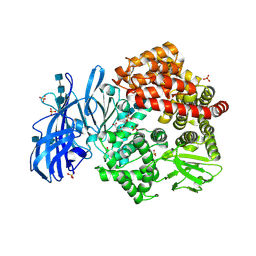 | | Crystal structure of porcine aminopeptidase-N with Aspartic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
7CE8
 
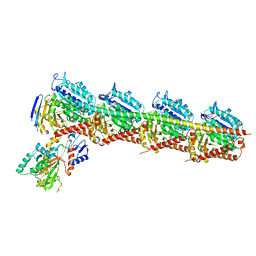 | | Crystal structure of T2R-TTL-Compound11 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.725 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
5F9W
 
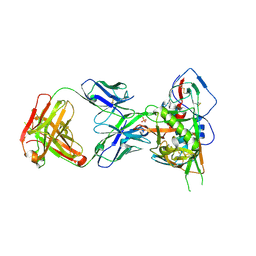 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of CH235-lineage antibody CH235, Light chain of CH235-lineage antibody CH235, ... | | Authors: | Chen, L, Zhou, T, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8911 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
7L7S
 
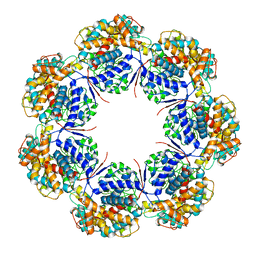 | | Human mitochondrial chaperonin mHsp60 | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Chen, L, Wang, J.C.Y. | | Deposit date: | 2020-12-30 | | Release date: | 2021-08-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the structural dynamics of human mitochondrial chaperonin mHsp60.
Sci Rep, 11, 2021
|
|
6JBH
 
 | | Cryo-EM structure and transport mechanism of a wall teichoic acid ABC transporter | | Descriptor: | TarG, TarH | | Authors: | Chen, L, Hou, W.T, Fan, T, Li, Y.H, Liu, B.H, Jiang, Y.L, Sun, L.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2019-01-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-electron Microscopy Structure and Transport Mechanism of a Wall Teichoic Acid ABC Transporter.
Mbio, 11, 2020
|
|
7CDA
 
 | | Crystal structure of T2R-TTL-PAC complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.659 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
7CEK
 
 | | Crystal structure of T2R-TTL-BML-284 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
7CE6
 
 | | Crystal structure of T2R-TTL-Compound9 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Small Molecules Promote Selective Denaturation and Degradation of Tubulin Heterodimers through a Low-Barrier Hydrogen Bond.
J.Med.Chem., 65, 2022
|
|
7CLD
 
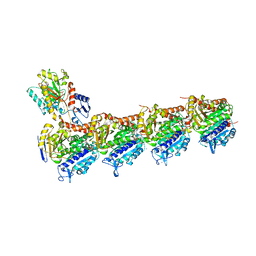 | | Crystal structure of T2R-TTL-Cevipabulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[2,6-bis(fluoranyl)-4-[3-(methylamino)propoxy]phenyl]-5-chloranyl-N-[(2S)-1,1,1-tris(fluoranyl)propan-2-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, CALCIUM ION, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Sci Adv, 7, 2021
|
|
7DP8
 
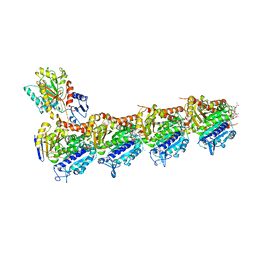 | | Crystal structure of T2R-TTL-Cevipabulin-eribulin complex | | Descriptor: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[2,6-bis(fluoranyl)-4-[3-(methylamino)propoxy]phenyl]-5-chloranyl-N-[(2S)-1,1,1-tris(fluoranyl)propan-2-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-12-18 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Sci Adv, 7, 2021
|
|
3H4J
 
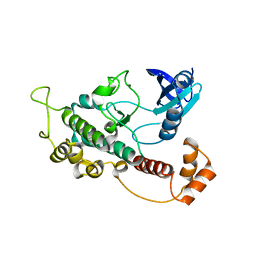 | | crystal structure of pombe AMPK KDAID fragment | | Descriptor: | SNF1-like protein kinase ssp2 | | Authors: | Chen, L, Jiao, Z.-H, Zheng, L.-S, Zhang, Y.-Y, Xie, S.-T, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the autoinhibition mechanism of AMP-activated protein kinase
Nature, 459, 2009
|
|
