7WWE
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 in an apo form | | Descriptor: | Phosphatidylinositol transfer protein CSR1 | | Authors: | Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2022-02-12 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7WVT
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh2 complexed with phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein CSR1 | | Authors: | Chen, L, Tan, L, Im, Y.J. | | Deposit date: | 2022-02-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of ligand recognition and transport by Sfh2, a yeast phosphatidylinositol transfer protein of the Sec14 superfamily.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6JT2
 
 | | Structure of human soluble guanylate cyclase in the NO activated state | | Descriptor: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | Authors: | Chen, L, Kang, Y, Liu, R, Wu, J.-X. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the mechanism of human soluble guanylate cyclase.
Nature, 574, 2019
|
|
6L65
 
 | | Sirtuin 2 protein with H3K18 myristoylated peptide | | Descriptor: | MYRISTIC ACID, NAD-dependent protein deacetylase sirtuin-2, PRO-ARG-LYS-GLN-LEU, ... | | Authors: | Chen, L.F. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sirtuin 2 protein with H3K18 myristoylated peptide
To Be Published
|
|
7E24
 
 | |
3LLP
 
 | | 1.8 Angstrom human fascin 1 crystal structure | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, Fascin, ... | | Authors: | Chen, L, Yang, S, Jakoncic, J, Zhang, J.J, Huang, X.-Y. | | Deposit date: | 2010-01-29 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Migrastatin analogues target fascin to block tumour metastasis.
Nature, 464, 2010
|
|
6L66
 
 | |
7E4T
 
 | | Human TRPC5 apo state structure at 3 angstrom | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CALCIUM ION, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2021-02-15 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7D4P
 
 | | Structure of human TRPC5 in complex with clemizole | | Descriptor: | (2S)-2-(hexadecanoyloxy)-3-hydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1-[(4-chlorophenyl)methyl]-2-(pyrrolidin-1-ylmethyl)benzimidazole, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7D4Q
 
 | | Structure of human TRPC5 in complex with HC-070 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 8-(3-chloranylphenoxy)-7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)purine-2,6-dione, CALCIUM ION, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
3JUA
 
 | |
6L72
 
 | | Sirtuin 2 demyristoylation native final product | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, [(2S,3R,4R,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2,4-bis(oxidanyl)oxolan-3-yl] tetradecanoate | | Authors: | Chen, L.F. | | Deposit date: | 2019-10-30 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Sirtuin 2 protein with H3K18 myristoylated peptide
To Be Published
|
|
6L71
 
 | |
3MQ0
 
 | | Crystal Structure of Agobacterium tumefaciens repressor BlcR | | Descriptor: | Transcriptional repressor of the blcABC operon | | Authors: | Chen, L. | | Deposit date: | 2010-04-27 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | The Agrobacterium tumefaciens Transcription Factor BlcR Is Regulated via Oligomerization.
J.Biol.Chem., 286, 2011
|
|
6JT0
 
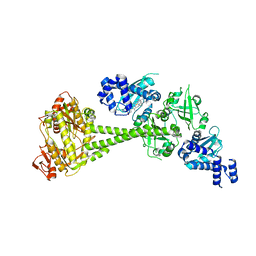 | | Structure of human soluble guanylate cyclase in the unliganded state | | Descriptor: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chen, L, Kang, Y, Liu, R, Wu, J.-X. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into the mechanism of human soluble guanylate cyclase.
Nature, 574, 2019
|
|
6JT1
 
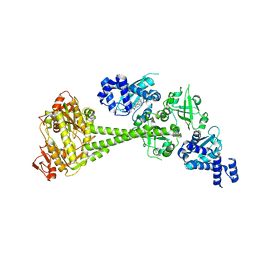 | | Structure of human soluble guanylate cyclase in the heme oxidised state | | Descriptor: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chen, L, Kang, Y, Liu, R, Wu, J.-X. | | Deposit date: | 2019-04-08 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into the mechanism of human soluble guanylate cyclase.
Nature, 574, 2019
|
|
7WIT
 
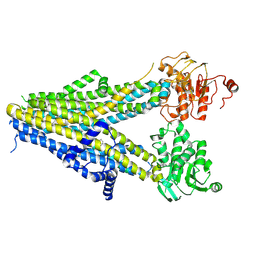 | | Structure of SUR1 in complex with mitiglinide | | Descriptor: | (2S)-4-[(3aR,7aS)-1,3,3a,4,5,6,7,7a-octahydroisoindol-2-yl]-4-oxidanylidene-2-(phenylmethyl)butanoic acid, ADENOSINE-5'-TRIPHOSPHATE, ATP-sensitive inward rectifier potassium channel 11,ATP-binding cassette sub-family C member 8 isoform X1 | | Authors: | Chen, L, Wang, M.M. | | Deposit date: | 2022-01-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural Insights Into the High Selectivity of the Anti-Diabetic Drug Mitiglinide
Front Pharmacol, 13, 2022
|
|
7WMV
 
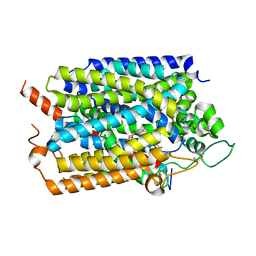 | | Structure of human SGLT1-MAP17 complex bound with LX2761 | | Descriptor: | N-[2-(dimethylamino)ethyl]-2-methyl-2-[4-[4-[[2-methyl-5-[(2S,3R,4R,5S,6R)-6-methylsulfanyl-3,4,5-tris(oxidanyl)oxan-2-yl]phenyl]methyl]phenyl]butanoylamino]propanamide, PDZK1-interacting protein 1, Sodium/glucose cotransporter 1 | | Authors: | Chen, L, Niu, Y, Cui, W. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mechanism of SGLT1 inhibitors.
Nat Commun, 13, 2022
|
|
7Y1J
 
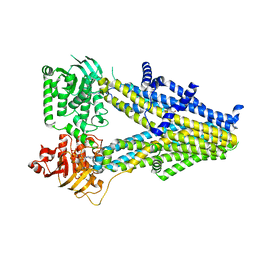 | | Structure of SUR2A in complex with Mg-ATP and repaglinide in the inward-facing conformation. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 9, MAGNESIUM ION, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1L
 
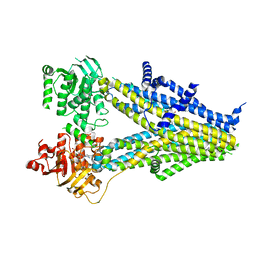 | | Structure of SUR2B in complex with Mg-ATP and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, MAGNESIUM ION, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1K
 
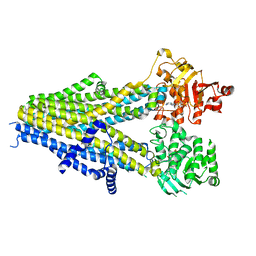 | | Structure of SUR2A in complex with Mg-ATP, Mg-ADP and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1M
 
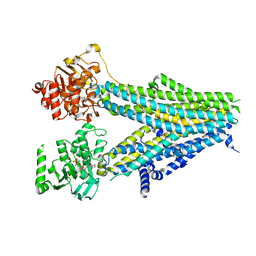 | | Structure of SUR2B in complex with Mg-ATP, Mg-ADP, and repaglinide in the inward-facing conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7Y1N
 
 | | Structure of SUR2B in complex with Mg-ATP, Mg-ADP, and repaglinide in the partially occluded state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Isoform SUR2B of ATP-binding cassette sub-family C member 9, ... | | Authors: | Chen, L, Ding, D, Hou, T. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | The inhibition mechanism of the SUR2A-containing K ATP channel by a regulatory helix.
Nat Commun, 14, 2023
|
|
7W7G
 
 | |
7YNJ
 
 | |
