2OC8
 
 | | Structure of Hepatitis C Viral NS3 protease domain complexed with NS4A peptide and ketoamide SCH503034 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C virus, ZINC ION, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R.S, Arasappan, A, Bennett, F, Bogen, S.L, Chen, K, Jao, E, Liu, Y.T, Lovey, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2KEM
 
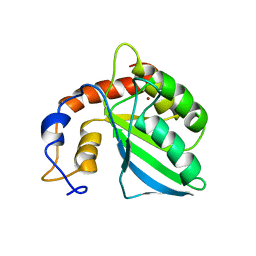 | | Extended structure of citidine deaminase domain of APOBEC3G | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Harjes, E, Gross, P.J, Chen, K, Lu, Y, Shindo, K, Nowarski, R, Gross, J.D, Kotler, M, Harris, R.S, Matsuo, H. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model
J.Mol.Biol., 389, 2009
|
|
1Q08
 
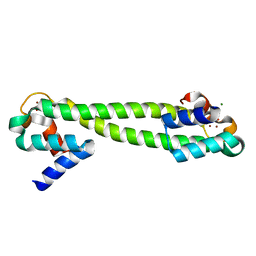 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator, at 1.9 A resolution (space group P212121) | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q06
 
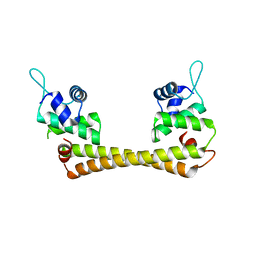 | | Crystal structure of the Ag(I) form of E. coli CueR, a copper efflux regulator | | Descriptor: | SILVER ION, Transcriptional regulator cueR | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q09
 
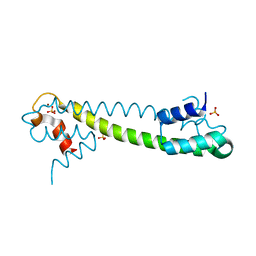 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group I4122) | | Descriptor: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | Authors: | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
7LHV
 
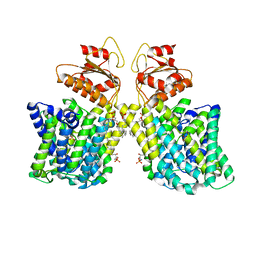 | | Structure of Arabidopsis thaliana sulfate transporter AtSULTR4;1 | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SULFATE ION, ... | | Authors: | Wang, L, Chen, K, Zhou, M. | | Deposit date: | 2021-01-26 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structure and function of an Arabidopsis thaliana sulfate transporter.
Nat Commun, 12, 2021
|
|
4P3P
 
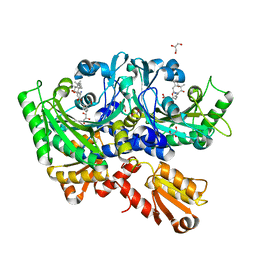 | | Structural Basis for Full-Spectrum Inhibition of Threonyl-tRNA Synthetase by Borrelidin 3 | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, GLYCEROL, Threonine--tRNA ligase, ... | | Authors: | Fang, P, Yu, X, Chen, K, Chen, X, Guo, M. | | Deposit date: | 2014-03-09 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase.
Nat Commun, 6, 2015
|
|
4P3O
 
 | | Structural Basis for Full-Spectrum Inhibition of Threonyl-tRNA Synthetase by Borrelidin 2 | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, GLYCEROL, Threonine--tRNA ligase, ... | | Authors: | Fang, P, Yu, X, Chen, K, Chen, X, Guo, M. | | Deposit date: | 2014-03-09 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase.
Nat Commun, 6, 2015
|
|
4P3N
 
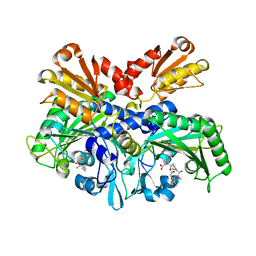 | | Structural Basis for Full-Spectrum Inhibition of Threonyl-tRNA Synthetase by Borrelidin 1 | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, Threonine--tRNA ligase, cytoplasmic, ... | | Authors: | Fang, P, Yu, X, Chen, K, Chen, X, Guo, M. | | Deposit date: | 2014-03-09 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for full-spectrum inhibition of translational functions on a tRNA synthetase.
Nat Commun, 6, 2015
|
|
3CVA
 
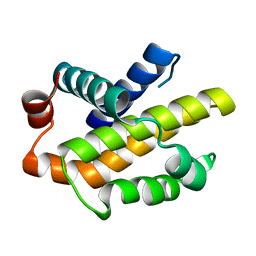 | | Human Bcl-xL containing a Trp to Ala mutation at position 137 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | Feng, Y, Zhang, L, Hu, T, Shen, X, Chen, K, Jiang, H, Liu, D. | | Deposit date: | 2008-04-18 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A conserved hydrophobic core at Bcl-x(L) mediates its structural stability and binding affinity with BH3-domain peptide of pro-apoptotic protein
Arch.Biochem.Biophys., 484, 2009
|
|
6M3R
 
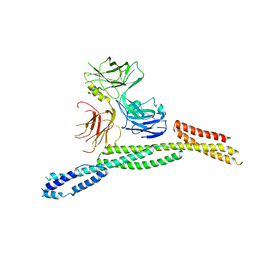 | | Crystal structure of AnkG/beta4-spectrin complex | | Descriptor: | Ankyrin-3, Spectrin beta chain | | Authors: | Li, J, Chen, K, Zhu, R, Zhang, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.313 Å) | | Cite: | Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
J.Mol.Biol., 432, 2020
|
|
6M3P
 
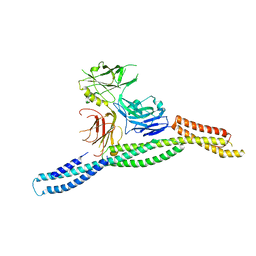 | | Crystal structure of AnkG/beta2-spectrin complex | | Descriptor: | Ankyrin-3, Spectrin beta chain, non-erythrocytic 1 | | Authors: | Li, J, Chen, K, Zhu, R, Zhang, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Structural Basis Underlying Strong Interactions between Ankyrins and Spectrins.
J.Mol.Biol., 432, 2020
|
|
6N64
 
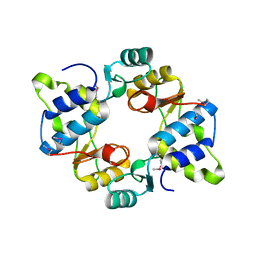 | | Crystal structure of mouse SMCHD1 hinge domain | | Descriptor: | Structural maintenance of chromosomes flexible hinge domain-containing protein 1, Uncharacterized peptide from Structural maintenance of chromosomes flexible hinge domain-containing protein 1 | | Authors: | Birkinshaw, R.W, Chen, K, Czabotar, P.E, Blewitt, M.E, Murphy, J.M. | | Deposit date: | 2018-11-25 | | Release date: | 2020-06-17 | | Last modified: | 2021-01-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the hinge domain of Smchd1 reveals its dimerization mode and nucleic acid-binding residues.
Sci.Signal., 13, 2020
|
|
1B0V
 
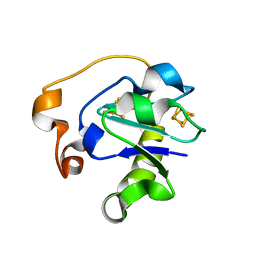 | | I40N MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN) | | Authors: | Sridhar, V, Prasad, G.S, Stout, C.D, Chen, K, Burgess, B.K. | | Deposit date: | 1998-11-12 | | Release date: | 2000-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Alteration of the reduction potential of the [4Fe-4S](2+/+) cluster of Azotobacter vinelandii ferredoxin I.
J.Biol.Chem., 274, 1999
|
|
1B0T
 
 | | D15K/K84D MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (FERREDOXIN I) | | Authors: | Sridhar, V, Stout, C.D, Chen, K, Kemper, M.A, Burgess, B.K. | | Deposit date: | 1998-11-12 | | Release date: | 1998-11-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the D15K/K84D Mutant of Azotobacter Vinelandii Ferredoxin I
To be Published
|
|
4JII
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Zopolrestat | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
4JIH
 
 | | Crystal Structure Of AKR1B10 Complexed With NADP+ And Epalrestat | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {5-[(2E)-2-methyl-3-phenylprop-2-en-1-ylidene]-4-oxo-2-thioxo-1,3-thiazolidin-3-yl}acetic acid | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
4JIR
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Epalrestat | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Zhang, L, Zheng, X, Zhang, H, Zhao, Y, Chen, K, Zhai, J, Hu, X. | | Deposit date: | 2013-03-06 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111).
Febs Lett., 587, 2013
|
|
5GZM
 
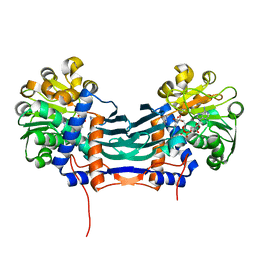 | | Cyclodeaminase_PA | | Descriptor: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ying, H, Chen, K. | | Deposit date: | 2016-09-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclodeaminase_PA
To Be Published
|
|
5GZL
 
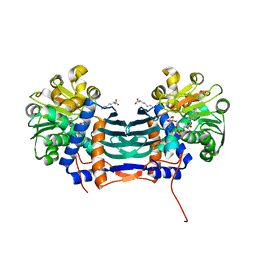 | | Cyclodeaminase_PA | | Descriptor: | LYSINE, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ying, H, Chen, K. | | Deposit date: | 2016-09-29 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclodeaminase_PA
To Be Published
|
|
5GZI
 
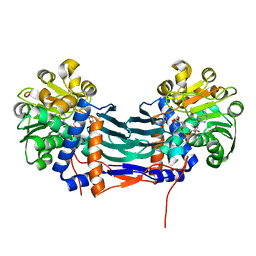 | | Cyclodeaminase_PA | | Descriptor: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ying, H, Chen, K. | | Deposit date: | 2016-09-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Cyclodeaminase_PA
To Be Published
|
|
5GZJ
 
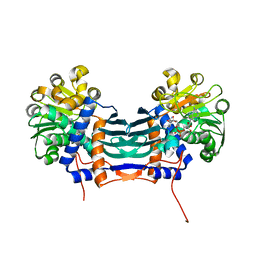 | | Cyclodeaminase_PA | | Descriptor: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ying, H, Chen, K. | | Deposit date: | 2016-09-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Cyclodeaminase_PA
To Be Published
|
|
5IE1
 
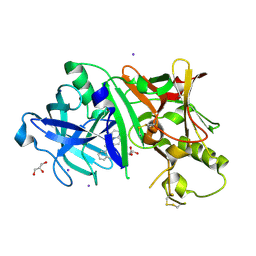 | | Crystal structure of BACE1 in complex with 3-(2-amino-6-(o-tolyl)quinolin-3-yl)-N-(3,3-dimethylbutyl)propanamide | | Descriptor: | 3-[2-amino-6-(2-methylphenyl)quinolin-3-yl]-N-(3,3-dimethylbutyl)propanamide, Beta-secretase 1, GLYCEROL, ... | | Authors: | Jordan, J.B, Whittington, D.A, Bartberger, M.D, Sickmier, E.A, Chen, K, Cheng, Y, Judd, T. | | Deposit date: | 2016-02-24 | | Release date: | 2016-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Fragment-Linking Approach Using (19)F NMR Spectroscopy To Obtain Highly Potent and Selective Inhibitors of beta-Secretase.
J.Med.Chem., 59, 2016
|
|
5HH9
 
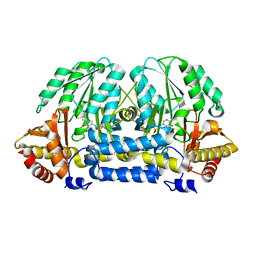 | | Structure of PvdN from Pseudomonas aeruginosa | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PvdN | | Authors: | Xia, H, Bai, G, Liu, S, Li, N, Xu, S, Chen, K, Yao, Q, Gu, L. | | Deposit date: | 2016-01-10 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Biochemical Characterization of a Novel L-Cystine Desulfurase involved in the biosynthesis of the major siderophore pyoverdine in Pseudomonas aeruginosa
To Be Published
|
|
2YKR
 
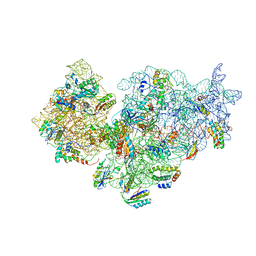 | | 30S ribosomal subunit with RsgA bound in the presence of GMPPNP | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Guo, Q, Yuan, Y, Xu, Y, Feng, B, Liu, L, Chen, K, Lei, J, Gao, N. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structural Basis for the Function of a Small Gtpase Rsga on the 30S Ribosomal Subunit Maturation Revealed by Cryoelectron Microscopy.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
