7KJV
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | HIV-1 viral RNA fragment, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
7KJW
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core with efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
6B19
 
 | | Architecture of HIV-1 reverse transcriptase initiation complex core | | Descriptor: | RNA genome fragment, reverse transcriptase p51 subunit, reverse transcriptase p66 subunit, ... | | Authors: | Larsen, K.P, Mathiharan, Y.K, Chen, D.H, Puglisi, J.D, Skiniotis, G, Puglisi, E.V. | | Deposit date: | 2017-09-18 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Architecture of an HIV-1 reverse transcriptase initiation complex.
Nature, 557, 2018
|
|
2I5N
 
 | | 1.96 A X-ray structure of photosynthetic reaction center from Rhodopseudomonas viridis:Crystals grown by microfluidic technique | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Li, L, Mustafi, D, Fu, Q, Tereshko, V, Chen, D.L, Tice, J.D, Ismagilov, R.F. | | Deposit date: | 2006-08-25 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Nanoliter microfluidic hybrid method for simultaneous screening and optimization validated with crystallization of membrane proteins.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4DSA
 
 | | Crystal Structure of DPP-IV with Compound C1 | | Descriptor: | 4-[({[(2R)-2-amino-3-(2,4,5-trifluorophenyl)propyl]sulfamoyl}amino)methyl]benzenesulfonamide, Dipeptidyl peptidase 4 | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2012-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal Structure of DPP-IV with Compound C1
To be Published
|
|
4FQ7
 
 | | Crystal structure of the maleate isomerase Iso from Pseudomonas putida S16 | | Descriptor: | Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
4FQ5
 
 | | Crystal structure of the maleate isomerase Iso(C200A) from Pseudomonas putida S16 with maleate | | Descriptor: | MALEIC ACID, Maleate cis-trans isomerase | | Authors: | Lu, Y, Chen, D, Zhang, Z, Li, Q, Wu, G, Xu, P. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside.
Mol.Microbiol., 87, 2013
|
|
8V2F
 
 | | Crystal structure of IRAK4 kinase domain with compound 9 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V1O
 
 | | Crystal structure of IRAK4 kinase domain with compound 4 | | Descriptor: | CHLORIDE ION, GLYCEROL, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-21 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
8V2L
 
 | | Crystal structure of IRAK4 kinase domain with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, Interleukin-1 receptor-associated kinase 4, N-{2-[4-(hydroxymethyl)phenyl]-6-(2-hydroxypropan-2-yl)-2H-indazol-5-yl}-6-(trifluoromethyl)pyridine-2-carboxamide | | Authors: | Weiss, M.M, Zheng, X, Browne, C.M, Campbell, V, Chen, D, Enerson, B, Fei, X, Huang, X, Klaus, C.R, Li, H, Mayo, M, McDonald, A.A, Paul, A, Sharma, K, Shi, Y, Slavin, A, Walter, D.M, Yuan, K, Zhang, Y, Zhu, X, Kelleher, J, Ji, N, Walker, D, Mainolfi, N. | | Deposit date: | 2023-11-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Discovery of KT-413, a Targeted Protein Degrader of IRAK4 and IMiD Substrates Targeting MYD88 Mutant Diffuse Large B-Cell Lymphoma.
J.Med.Chem., 67, 2024
|
|
5Y22
 
 | | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV | | Descriptor: | 22AA-PSTD peptide | | Authors: | Lu, B, Liao, S.M, Huang, J.M, Lu, Z.L, Chen, D, Liu, X.H, Zhou, G.P, Huang, R.B. | | Deposit date: | 2017-07-23 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV
To Be Published
|
|
4DSZ
 
 | | Crystal Structure of DPP-IV with Compound C2 | | Descriptor: | (2R)-4-[4-(3-methylphenyl)-1H-1,2,3-triazol-1-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine, Dipeptidyl peptidase 4 | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2012-02-20 | | Release date: | 2013-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of DPP-IV with Compound C2
To be Published
|
|
6AHZ
 
 | | The NMR Structure of the Polysialyltranseferase Domain (PSTD) in Polysialyltransferase ST8siaIV | | Descriptor: | CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase | | Authors: | Liu, X.H, Lu, B, Peng, L.X, Liao, S.M, Zhou, F, Chen, D, Lu, Z.L, Zhou, G.P, Huang, R.B. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Inhibition of Polysialyltranseferase ST8SiaIV Through Heparin Binding to Polysialyltransferase Domain (PSTD).
Med Chem, 15, 2019
|
|
7VH5
 
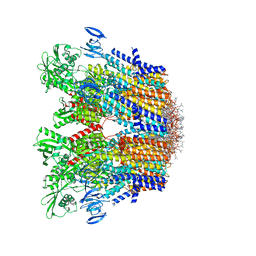 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the autoinhibited state (pH 7.4, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Plasma membrane ATPase 1, SPHINGOSINE | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
7VH6
 
 | | Cryo-EM structure of the hexameric plasma membrane H+-ATPase in the active state (pH 6.0, BeF3-, conformation 1, C1 symmetry) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, BERYLLIUM TRIFLUORIDE ION, Plasma membrane ATPase 1 | | Authors: | Zhao, P, Zhao, C, Chen, D, Yun, C, Li, H, Bai, L. | | Deposit date: | 2021-09-21 | | Release date: | 2021-11-24 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and activation mechanism of the hexameric plasma membrane H + -ATPase.
Nat Commun, 12, 2021
|
|
5YFM
 
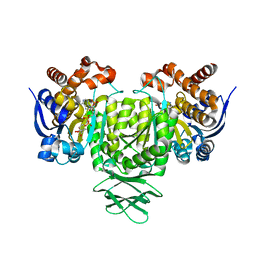 | |
5YFN
 
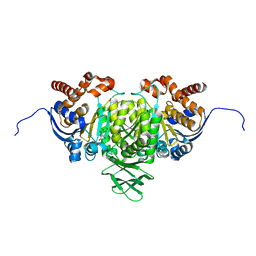 | | Human isocitrate dehydrogenase 1 bound with isocitrate | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, MAGNESIUM ION, ... | | Authors: | Nordlund, P, Chen, D, Jansson, A, Larsson, A. | | Deposit date: | 2017-09-21 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human isocitrate dehydrogenase 1 bound with isocitrate
To Be Published
|
|
4J3J
 
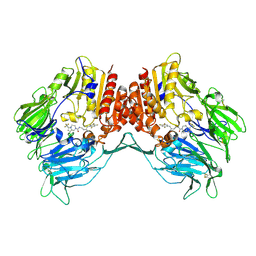 | | Crystal Structure of DPP-IV with Compound C3 | | Descriptor: | Dipeptidyl peptidase 4, N-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butyl]-6-(trifluoromethyl)-3,4-dihydropyrrolo[1,2-a]pyrazine-2(1H)-carboxamide | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 4-(2,4,5-trifluorophenyl)butane-1,3-diamines as dipeptidyl peptidase IV inhibitors
Chemmedchem, 8, 2013
|
|
5Y3U
 
 | | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV in the Presence of Polysialic Acid (PolySia) | | Descriptor: | PSTD-22AA-PolySia | | Authors: | Liao, S.M, Liu, X.H, Lu, B, Peng, L.X, Chen, D, Huang, R.B, Zhou, G.P. | | Deposit date: | 2017-07-31 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR-Based Model of the 22 Amino Acid Peptide in Polysialyltransferase Domain (PSTD) of the Polysialyltransferase ST8Sia IV in the Presence of Polysialic Acid (PolySia)
To Be Published
|
|
7DLZ
 
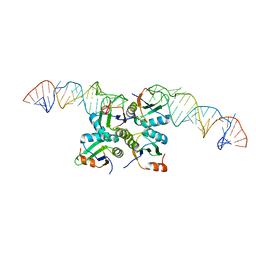 | | Crystal Structure of Methyltransferase Ribozyme | | Descriptor: | RNA (45-MER), U1 small nuclear ribonucleoprotein A | | Authors: | Gan, J.H, Gao, Y.Q, Jiang, H.Y, Chen, D.R, Murchie, A.I.H. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
7DWH
 
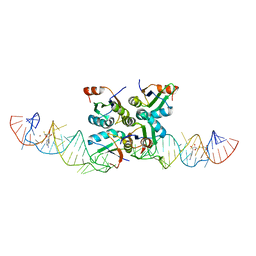 | | Complex structure of SAM-dependent methyltransferase ribozyme | | Descriptor: | COPPER (II) ION, RNA (45-MER), S-ADENOSYLMETHIONINE, ... | | Authors: | Jiang, H.Y, Gao, Y.Q, Chen, D.R, Murchie, A. | | Deposit date: | 2021-01-17 | | Release date: | 2021-10-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
Nat Catal, 4, 2021
|
|
4GJH
 
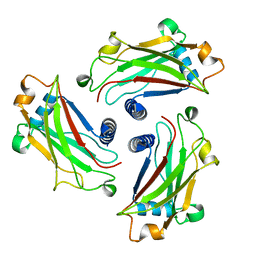 | | Crystal Structure of the TRAF domain of TRAF5 | | Descriptor: | TNF receptor-associated factor 5 | | Authors: | Zhang, P, Reichardt, A, Liang, H, Wang, Y, Cheng, D, Aliyari, R, Cheng, G, Liu, Y. | | Deposit date: | 2012-08-09 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Single Amino Acid Substitutions Confer the Antiviral Activity of the TRAF3 Adaptor Protein onto TRAF5
Sci.Signal., 5, 2012
|
|
8IMZ
 
 | | Cryo-EM structure of mouse Piezo1-MDFIC complex (composite map) | | Descriptor: | MyoD family inhibitor domain-containing protein, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Zhou, Z, Ma, X, Lin, Y, Cheng, D, Bavi, N, Li, J.V, Sutton, D, Yao, M, Harvey, N, Corry, B, Zhang, Y, Cox, C.D. | | Deposit date: | 2023-03-07 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | MyoD-family inhibitor proteins act as auxiliary subunits of Piezo channels.
Science, 381, 2023
|
|
7LXQ
 
 | |
7LXP
 
 | |
