1T3L
 
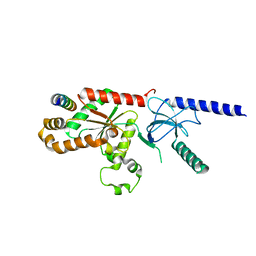 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core in Complex with Alpha1 Interaction Domain | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, Voltage-dependent L-type calcium channel alpha-1S subunit | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Voltage-Dependent Calcium Channel Beta Subunit Functional Core and Its Complex with the Alpha1 Interaction Domain
NEURON, 42, 2004
|
|
9JDC
 
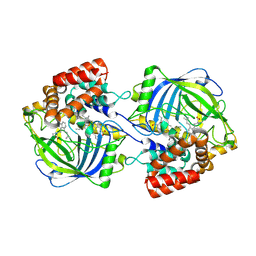 | | Structure of chanoclavine synthase from Claviceps fusiformis in complex with prechanoclavine | | Descriptor: | (2~{S})-2-(methylamino)-3-[4-[(1~{E})-3-methylbuta-1,3-dienyl]-1~{H}-indol-3-yl]propanoic acid, Catalase easC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, Z.W, Wang, T, Li, X, Shen, P.P, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-08-31 | | Release date: | 2025-01-01 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Chanoclavine synthase operates by an NADPH-independent superoxide mechanism.
Nature, 2025
|
|
9JDB
 
 | | Structure of chanoclavine synthase from Claviceps fusiformis | | Descriptor: | Catalase easC, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Liu, Z.W, Wang, T, Li, X, Shen, P.P, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-08-31 | | Release date: | 2025-01-01 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Chanoclavine synthase operates by an NADPH-independent superoxide mechanism.
Nature, 2025
|
|
9JN6
 
 | | Crystal structure of KtzT-C197A in complex with HEME and product | | Descriptor: | (3~{S})-1,2-diazinane-3-carboxylic acid, FMN-binding protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, Y.L, Yang, Y.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural Insights into the N-N Bond-Formation Mechanism of the Heme-Dependent Piperazate Synthase KtzT
Acs Catalysis, 15, 2025
|
|
9JN4
 
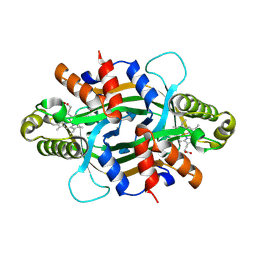 | | Crystal structure of KtzT-C197A in complex with HEME | | Descriptor: | FMN-binding protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, Y.L, Yang, Y.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Insights into the N-N Bond-Formation Mechanism of the Heme-Dependent Piperazate Synthase KtzT
Acs Catalysis, 15, 2025
|
|
9JN5
 
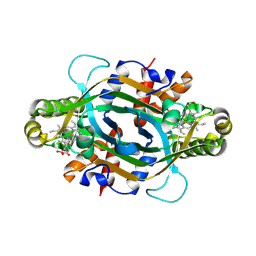 | | Crystal structure of KtzT-C197A in complex with HEME and substrate | | Descriptor: | FMN-binding protein, N~5~-hydroxy-L-ornithine, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, Y.L, Yang, Y.Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2024-09-22 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the N-N Bond-Formation Mechanism of the Heme-Dependent Piperazate Synthase KtzT
Acs Catalysis, 15, 2025
|
|
4LXL
 
 | | Crystal structure of JMJD2B complexed with pyridine-2,4-dicarboxylic acid and H3K9me3 | | Descriptor: | H3 peptide, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | Wang, W.-C, Chu, C.-H, Chen, C.-C. | | Deposit date: | 2013-07-30 | | Release date: | 2014-07-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of JMJD2B complexed with pyridine-2,4-dicarboxylic acid and H3K9me3
To be Published
|
|
5B01
 
 | |
1T3S
 
 | | Structural Analysis of the Voltage-Dependent Calcium Channel Beta Subunit Functional Core | | Descriptor: | Dihydropyridine-sensitive L-type, calcium channel beta-2 subunit, MERCURY (II) ION | | Authors: | Opatowsky, Y, Chen, C.-C, Campbell, K.P, Hirsch, J.A. | | Deposit date: | 2004-04-27 | | Release date: | 2004-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain.
Neuron, 42, 2004
|
|
5B0J
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-undecyl maltoside | | Descriptor: | MoeN5,DNA-binding protein 7d, UNDECYL-MALTOSIDE | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B0M
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-dodecyl maltoside | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B00
 
 | | Structure of the prenyltransferase MoeN5 in complex with geranyl pyrophosphate | | Descriptor: | GERANYL DIPHOSPHATE, MoeN5 | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B0L
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-nonyl glucoside | | Descriptor: | MoeN5,DNA-binding protein 7d, nonyl beta-D-glucopyranoside | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B03
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with geranyl pyrophosphate | | Descriptor: | GERANYL DIPHOSPHATE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B0K
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-decyl maltoside | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B0I
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with beta-octyl glucoside | | Descriptor: | MoeN5,DNA-binding protein 7d, octyl beta-D-glucopyranoside | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-30 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5B02
 
 | | Structure of the prenyltransferase MoeN5 with a fusion protein tag of Sso7d | | Descriptor: | MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Zhang, L, Chen, C.-C, Guo, R.-T, Oldfield, E.O. | | Deposit date: | 2015-10-27 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Moenomycin Biosynthesis: Structure and Mechanism of Action of the Prenyltransferase MoeN5.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
8JMO
 
 | | Structure of a leaf-branch compost cutinase, ICCG in complex with 4-((4-Hydroxybutoxy)carbonyl)benzoic acid | | Descriptor: | 4-(4-oxidanylbutoxycarbonyl)benzoic acid, CALCIUM ION, Leaf-branch compost cutinase | | Authors: | Yang, Y, Xue, T, Zheng, Y, Cheng, S, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Remodeling the polymer-binding cavity to improve the efficacy of PBAT-degrading enzyme.
J Hazard Mater, 464, 2023
|
|
8JMP
 
 | | Structure of a leaf-branch compost cutinase, ICCG in complex with 1,4-butanediol terephthalate | | Descriptor: | 4-[4-(4-carboxyphenyl)carbonyloxybutoxycarbonyl]benzoic acid, CALCIUM ION, Leaf-branch compost cutinase | | Authors: | Yang, Y, Xue, T, Zheng, Y, Cheng, S, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2023-06-05 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Remodeling the polymer-binding cavity to improve the efficacy of PBAT-degrading enzyme.
J Hazard Mater, 464, 2023
|
|
5GWV
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with a substrate analogue | | Descriptor: | (2R)-3-dimethoxyphosphoryloxy-2-[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trienoxy]propanoic acid, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.-P, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
5GWW
 
 | | Structure of MoeN5-Sso7d fusion protein in complex with a permethylated substrate analogue | | Descriptor: | MoeN5,DNA-binding protein 7d, methyl (2R)-3-dimethoxyphosphoryloxy-2-[(2Z,6E)-3,7,11-trimethyldodeca-2,6,10-trienoxy]propanoate | | Authors: | Ko, T.-P, Guo, R.-T, Chen, C.-C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
8H7P
 
 | | Crystal structure of aqualigase bound with Suc-AAPF | | Descriptor: | 5,6-DIHYDRO-BENZO[H]CINNOLIN-3-YLAMINE, CALCIUM ION, Subtilisin, ... | | Authors: | Li, H, Ma, M.Z, Zhang, L.J, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of aqualigase bound with Suc-AAPF
To Be Published
|
|
8H7O
 
 | | Crystal structure of aqualigase | | Descriptor: | CALCIUM ION, ISOPROPYL ALCOHOL, Subtilisin | | Authors: | Li, H, Ma, M.Z, Zhang, L.J, Dai, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-10-20 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of aqualigase
To Be Published
|
|
7BR2
 
 | |
8HJT
 
 | | Crystal Structure of Intracellular B30.2 Domain of VpBTN3 and VpBTN2 in Complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Butyrophylin 3, ... | | Authors: | Yang, Y.Y, Shen, P.P, Li, X, Yi, S.M, Zhang, M.T, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-11-23 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
