8IHS
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHR
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 in complex with Phe | | Descriptor: | Amidohydrolase family protein, PHENYLALANINE, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8IHQ
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 | | Descriptor: | Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-02-23 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8J85
 
 | | Cryo-EM structure of ochratoxin A-detoxifying amidohydrolase ADH3 mutant S88E in complex with ochratoxin A | | Descriptor: | (2~{S})-2-[[(3~{R})-5-chloranyl-3-methyl-8-oxidanyl-1-oxidanylidene-3,4-dihydroisochromen-7-yl]carbonylamino]-3-phenyl-propanoic acid, Amidohydrolase family protein, ZINC ION | | Authors: | Dai, L.H, Niu, D, Huang, J.-W, Li, X, Shen, P.P, Li, H, Hu, Y.M, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase.
J Hazard Mater, 458, 2023
|
|
8JI5
 
 | | Crystal structure of AetD in complex with 5-bromo-L-tryptophan and two Fe2+ | | Descriptor: | 5-bromo-L-tryptophan, AetD, FE (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI3
 
 | | Crystal structure of AetD in complex with 5,7-dibromo-L-tryptophan and two Fe2+ | | Descriptor: | (2S)-2-azanyl-3-[5,7-bis(bromanyl)-1H-indol-3-yl]propanoic acid, AetD, FE (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI6
 
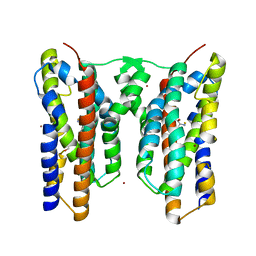 | | Crystal structure of AetD in complex with L-tryptophan | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AetD, FE (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI2
 
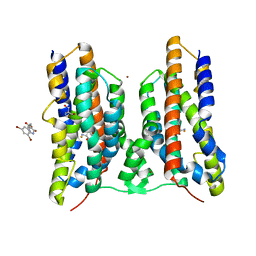 | | Crystal structure of AetD in complex with 5,7-dibromo-L-tryptophan | | Descriptor: | (2S)-2-azanyl-3-[5,7-bis(bromanyl)-1H-indol-3-yl]propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AetD, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI4
 
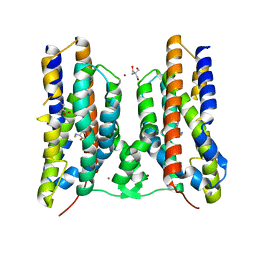 | | Crystal structure of AetD in complex with 5-bromo-L-tryptophan | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-bromo-L-tryptophan, AetD, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8JI7
 
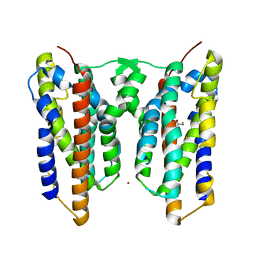 | | Crystal structure of AetD in complex with L-tryptophan and two Fe2+ | | Descriptor: | AetD, FE (II) ION, NICKEL (II) ION, ... | | Authors: | Li, H, Dai, L, Zheng, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-05-26 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The structural and functional investigation into an unusual nitrile synthase.
Nat Commun, 14, 2023
|
|
8J45
 
 | | Crystal structure of a Pichia pastoris-expressed IsPETase variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poly(ethylene terephthalate) hydrolase | | Authors: | Li, X, He, H.L, Long, X, Niu, D, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-04-19 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Complete decomposition of poly(ethylene terephthalate) by crude PET hydrolytic enzyme produced in Pichia pastoris
Chem Eng J, 2023
|
|
7Y9M
 
 | | Crystal structure of P450 BM3-2F from Bacillus megaterium | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of P450 BM3-2F from Bacillus megaterium
to be published
|
|
7Y9L
 
 | | Crystal structure of P450 BM3-2F from Bacillus megaterium in complex with 2-Hydroxy-5-Nitrobenzonitrile | | Descriptor: | 5-nitro-2-oxidanyl-benzenecarbonitrile, Bifunctional cytochrome P450/NADPH--P450 reductase, NICKEL (II) ION, ... | | Authors: | Wang, Q, Zhang, L.L, Liu, W.D, Huang, J.-W, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of P450 BM3-2F from Bacillus megaterium in complex with 2-Hydroxy-5-Nitrobenzonitrile
to be published
|
|
7EWA
 
 | | GDP-bound KRAS G12D in complex with TH-Z827 | | Descriptor: | 4-[(1~{R},5~{S})-3,8-diazabicyclo[3.2.1]octan-8-yl]-7-(8-methylnaphthalen-1-yl)-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
7EW9
 
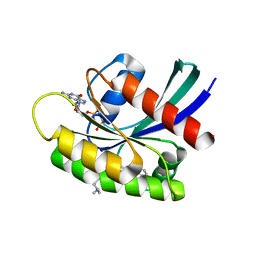 | | GDP-bound KRAS G12D in complex with TH-Z816 | | Descriptor: | 7-(8-methylnaphthalen-1-yl)-4-[(2~{R})-2-methylpiperazin-1-yl]-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
7EWB
 
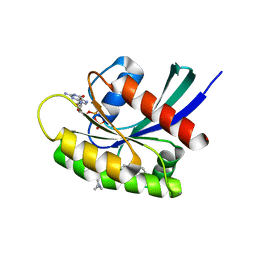 | | GDP-bound KRAS G12D in complex with TH-Z835 | | Descriptor: | 4-[(1~{S},5~{R})-3,8-diazabicyclo[3.2.1]octan-3-yl]-7-(8-methylnaphthalen-1-yl)-2-[[(2~{S})-1-methylpyrrolidin-2-yl]methoxy]-6,8-dihydro-5~{H}-pyrido[3,4-d]pyrimidine, GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, ... | | Authors: | Shen, P, Yang, Y, Yang, Y, Zhang, L, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2021-05-25 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | KRAS(G12D) can be targeted by potent inhibitors via formation of salt bridge.
Cell Discov, 8, 2022
|
|
7FIR
 
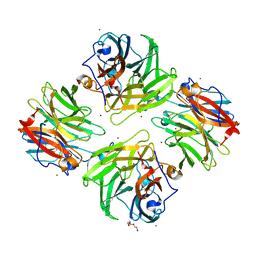 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with 1,4-mannobiose | | Descriptor: | Beta-1,2-mannobiose phosphorylase, PENTAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIQ
 
 | | The crystal structure of mannose-bound beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIP
 
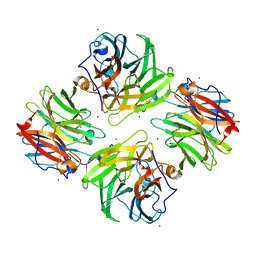 | | The native structure of beta-1,2-mannobiose phosphorylase from Thermoanaerobacter sp. | | Descriptor: | Beta-1,2-mannobiose phosphorylase, ZINC ION | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7FIS
 
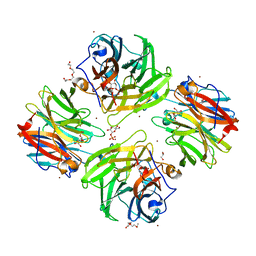 | | The crystal structure of beta-1,2-mannobiose phosphorylase in complex with mannose 1-phosphate (M1P) | | Descriptor: | 1-O-phosphono-alpha-D-mannopyranose, Beta-1,2-mannobiose phosphorylase, GLYCEROL, ... | | Authors: | Dai, L, Chang, Z, Yang, J, Liu, W, Yang, Y, Chen, C.-C, Zhang, L, Huang, J, Sun, Y, Guo, R.-T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural investigation of a thermostable 1,2-beta-mannobiose phosphorylase from Thermoanaerobacter sp. X-514.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
7F7M
 
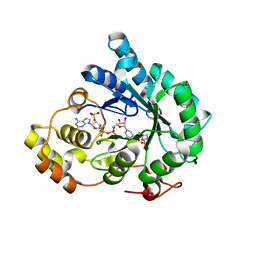 | | AKR4C17 in complex with NADP+ and glyphosate | | Descriptor: | AKR4-2, GLYPHOSATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7F7L
 
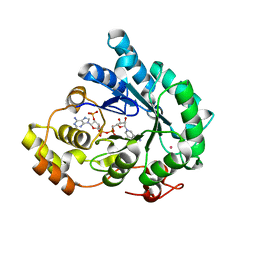 | | Crystal structure of AKR4C17 bound with NADPH | | Descriptor: | AKR4-2, COBALT (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7F7K
 
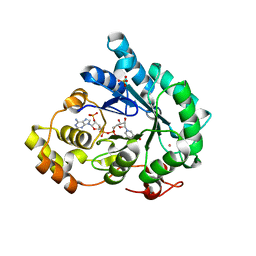 | | Crystal structure of AKR4C17 bound with NADP+ | | Descriptor: | AKR4-2, COBALT (II) ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7F7J
 
 | | The crystal structure of AKR4C17 | | Descriptor: | AKR4-2, COBALT (II) ION, SULFATE ION | | Authors: | Li, H, Yang, Y, Hu, Y, Chen, C.-C, Huang, J.-W, Min, J, Dai, L, Guo, R.-T. | | Deposit date: | 2021-06-30 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural analysis and engineering of aldo-keto reductase from glyphosate-resistant Echinochloa colona
J Hazard Mater, 436, 2022
|
|
7Y9O
 
 | | Crystal structure of a CYP109B4 variant from Bacillus sonorensis | | Descriptor: | CALCIUM ION, Cytochrome P450 monooxygenase YjiB, IMIDAZOLE, ... | | Authors: | Shen, P.P, Huang, J.-W, Li, X, Liu, W.D, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Rationally Controlling Selective Steroid Hydroxylation via Scaffold Sampling of a P450 Family
Acs Catalysis, 13, 2023
|
|
