3RF6
 
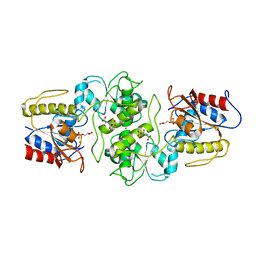 | | Crystal structure of glycerol-3 phosphate bound HAD-like phosphatase from Saccharomyces cerevisiae | | Descriptor: | CITRATE ANION, MAGNESIUM ION, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Nocek, B, Kuznetsova, K, Evdokimova, E, Savchenko, A, Iakunine, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-05 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of glycerol-3 phosphate bound HAD-like phosphatase from Saccharomyces cerevisiae
TO BE PUBLISHED
|
|
3CQV
 
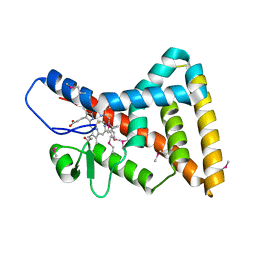 | | Crystal structure of Reverb beta in complex with heme | | Descriptor: | Nuclear receptor subfamily 1 group D member 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xu, X, Dong, A, Pardee, K.I, Reinking, J, Krause, H, Schuetz, A, Zhang, R, Cui, H, Edwards, A, Arrowsmith, C.H, Weigelt, J, Bountra, C, Savchenko, A, Botchkarev, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-03 | | Release date: | 2008-08-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta.
Plos Biol., 7, 2009
|
|
3K29
 
 | | Structure of a putative YscO homolog CT670 from Chlamydia trachomatis | | Descriptor: | Putative uncharacterized protein | | Authors: | Lam, R, Singer, A, Skarina, T, Onopriyenko, O, Bochkarev, A, Brunzelle, J.S, Edwards, A.M, Anderson, W.F, Chirgadze, N.Y, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and protein-protein interaction studies on Chlamydia trachomatis protein CT670 (YscO Homolog).
J.Bacteriol., 192, 2010
|
|
1N1O
 
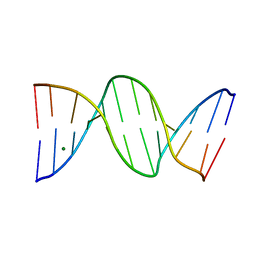 | | Crystal Structure of a B-form DNA Duplex Containing (L)-alpha-threofuranosyl (3'-2') Nucleosides: A Four-Carbon Sugar is Easily Accommodated into the Backbone of DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(TFT)P*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Wilds, C.J, Wawrzak, Z, Krishnamurthy, R, Eschenmoser, A, Egli, M. | | Deposit date: | 2002-10-18 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a B-Form DNA Duplex Containing (L)-alpha-Threofuranosyl (3'-->2') Nucleosides: A
Four-Carbon Sugar Is Easily Accommodated into the Backbone of DNA
J.Am.Chem.Soc., 124, 2002
|
|
4RXV
 
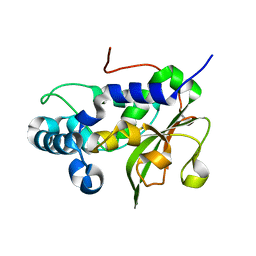 | | The crystal structure of the N-terminal fragment of uncharacterized protein from Legionella pneumophila | | Descriptor: | hypothetical protein lpg0944 | | Authors: | Nocek, B, Cuff, M, Evdokimova, E, Egorova, O, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-12 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
3TYP
 
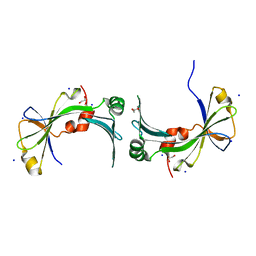 | | The crystal structure of the inorganic triphosphatase NE1496 | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Uncharacterized protein | | Authors: | Lunin, V.V, Skarina, T, Onopriyenko, O, Binkowski, T.A, Joachimiak, A, Edwards, A.M, Savchenko, A. | | Deposit date: | 2011-09-26 | | Release date: | 2012-05-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A specific inorganic triphosphatase from Nitrosomonas europaea: structure and catalytic mechanism.
J.Biol.Chem., 286, 2011
|
|
3QVM
 
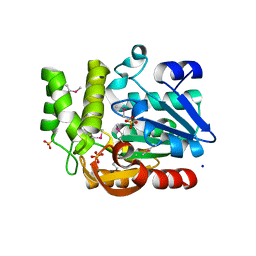 | | The structure of olei00960, a hydrolase from Oleispira antarctica | | Descriptor: | CALCIUM ION, CHLORIDE ION, Olei00960, ... | | Authors: | Singer, A.U, Kagan, O, Kim, Y, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-04-13 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
6RZ4
 
 | | Crystal structure of cysteinyl leukotriene receptor 1 in complex with pranlukast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Cysteinyl leukotriene receptor 1,Soluble cytochrome b562,Cysteinyl leukotriene receptor 1, OLEIC ACID, ... | | Authors: | Luginina, A, Gusach, A, Marin, E, Mishin, A, Brouillette, R, Popov, P, Shiryaeva, A, Besserer-Offroy, E, Longpre, J.M, Lyapina, E, Ishchenko, A, Patel, N, Polovinkin, V, Safronova, N, Bogorodskiy, A, Edelweiss, E, Liu, W, Batyuk, A, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs.
Sci Adv, 5, 2019
|
|
3UDO
 
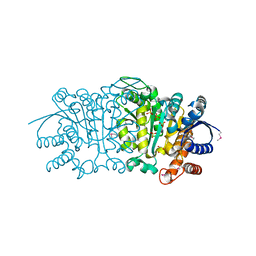 | | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, SULFATE ION | | Authors: | Tkaczuk, K.L, Chruszcz, M, Blus, B.J, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-28 | | Release date: | 2011-11-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of putative isopropylamlate dehydrogenase from Campylobacter jejuni
To be Published
|
|
3K6C
 
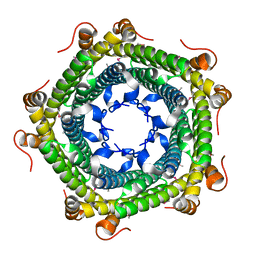 | | Crystal structure of protein ne0167 from nitrosomonas europaea | | Descriptor: | Uncharacterized protein NE0167 | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Protein Ne0167 from Nitrosomonas Europaea
To be Published
|
|
3UUW
 
 | | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Kudritska, M, Grimshaw, S, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-28 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | 1.63 Angstrom Resolution Crystal Structure of Dehydrogenase (MviM) from Clostridium difficile.
TO BE PUBLISHED
|
|
5UNF
 
 | | XFEL structure of human angiotensin II type 2 receptor (Monoclinic form) in complex with compound 1 (N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]) | | Descriptor: | Chimera protein of Type-2 angiotensin II receptor and Soluble cytochrome b562, N-benzyl-N-(2-ethyl-4-oxo-3-{[2'-(2H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydroquinazolin-6-yl)thiophene-2-carboxamide | | Authors: | Zhang, H, Han, G.W, Batyuk, A, Ishchenko, A, White, K.L, Patel, N, Sadybekov, A, Zamlynny, B, Rudd, M.T, Hollenstein, K, Tolstikova, A, White, T.A, Hunter, M.S, Weierstall, U, Liu, W, Babaoglu, K, Moore, E.L, Katz, R.D, Shipman, J.M, Garcia-Calvo, M, Sharma, S, Sheth, P, Soisson, S.M, Stevens, R.C, Katritch, V, Cherezov, V. | | Deposit date: | 2017-01-30 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for selectivity and diversity in angiotensin II receptors.
Nature, 544, 2017
|
|
5L1A
 
 | | Crystal structure of uncharacterized protein LPG2271 from Legionella pneumophila | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Uncharacterized protein | | Authors: | Chang, C, Xu, X, Cui, H, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-07-28 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of uncharacterized protein LPG2271 from Legionella pneumophila
To Be Published
|
|
3UWD
 
 | | Crystal Structure of Phosphoglycerate Kinase from Bacillus Anthracis | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Zheng, H, Chruszcz, M, Porebski, P, Kudritska, M, Grimshaw, S, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-01 | | Release date: | 2012-01-11 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structures of putative phosphoglycerate kinases from B. anthracis and C. jejuni.
J.Struct.Funct.Genom., 13, 2012
|
|
3UMC
 
 | | Crystal Structure of the L-2-Haloacid Dehalogenase PA0810 | | Descriptor: | CHLORIDE ION, SODIUM ION, haloacid dehalogenase | | Authors: | Petit, P, Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-13 | | Release date: | 2012-11-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
6RZ5
 
 | | XFEL crystal structure of the human cysteinyl leukotriene receptor 1 in complex with zafirlukast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Cysteinyl leukotriene receptor 1,Soluble cytochrome b562,Cysteinyl leukotriene receptor 1, OLEIC ACID, ... | | Authors: | Luginina, A, Gusach, A, Marin, E, Mishin, A, Brouillette, R, Popov, P, Shiryaeva, A, Besserer-Offroy, E, Longpre, J.M, Lyapina, E, Ishchenko, A, Patel, N, Polovinkin, V, Safronova, N, Bogorodskiy, A, Edelweiss, E, Liu, W, Batyuk, A, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-based mechanism of cysteinyl leukotriene receptor inhibition by antiasthmatic drugs.
Sci Adv, 5, 2019
|
|
3GL5
 
 | | Crystal structure of probable DsbA oxidoreductase SCO1869 from Streptomyces coelicolor | | Descriptor: | ACETATE ION, Putative DsbA oxidoreductase SCO1869, SODIUM ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of probable DsbA oxidoreductase SCO1869 from Streptomyces coelicolor
To be Published
|
|
3OBB
 
 | | Crystal structure of a possible 3-hydroxyisobutyrate Dehydrogenase from pseudomonas aeruginosa pao1 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tan, K, Singer, A.U, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Yakunin, A.F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-06 | | Release date: | 2010-08-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Studies of Uncharacterized Protein PA0743 from Pseudomonas aeruginosa Revealed NAD+-dependent L-Serine Dehydrogenase.
J.Biol.Chem., 287, 2012
|
|
5L0L
 
 | | Crystal structure of Uncharacterized protein LPG0439 | | Descriptor: | CHLORIDE ION, SULFATE ION, Uncharacterized protein | | Authors: | Chang, C, Skarina, T, Khutoreskaya, G, Savchenko, A, Joachimiak, A. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Uncharacterized protein LPG0439
To Be Published
|
|
3QGM
 
 | | p-nitrophenyl phosphatase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, p-nitrophenyl phosphatase (Pho2) | | Authors: | Osipiuk, J, Zheng, H, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-01-24 | | Release date: | 2011-02-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
4LAT
 
 | | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A in complex with phosphate | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHATE ION, Phosphate-binding protein PstS 1 | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A in complex with phosphate
To be Published
|
|
6CZP
 
 | | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-09 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN.
To Be Published
|
|
3OMB
 
 | | Crystal structure of extracellular solute-binding protein from Bifidobacterium longum subsp. infantis | | Descriptor: | Extracellular solute-binding protein, family 1, MAGNESIUM ION | | Authors: | Chang, C, Xu, X, Chin, S, Cui, H, Dong, A, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of extracellular solute-binding protein from Bifidobacterium longum subsp. infantis
To be Published
|
|
3TPF
 
 | | Crystal structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Ornithine carbamoyltransferase | | Authors: | Shabalin, I.G, Onopriyenko, O, Grimshaw, S, Porebski, P.J, Grabowski, M, Savchenko, A, Chruszcz, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of anabolic ornithine carbamoyltransferase from Campylobacter jejuni at 2.7 A resolution.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3ON1
 
 | | The structure of a protein with unknown function from Bacillus halodurans C | | Descriptor: | BH2414 protein | | Authors: | Fan, Y, Kagan, O, Savchenko, A, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of a protein with unknown function from Bacillus halodurans C
To be Published
|
|
