1EL9
 
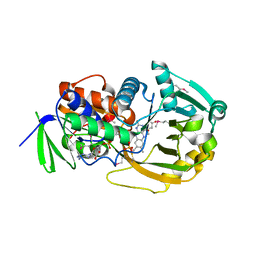 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR [METHYLTHIO]ACETATE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
2MLA
 
 | |
1EL8
 
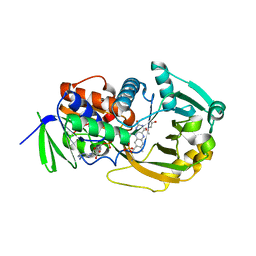 | | COMPLEX OF MONOMERIC SARCOSINE OXIDASE WITH THE INHIBITOR [METHYLSELENO]CETATE | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Wagner, M.A, Trickey, P, Chen, Z.-W, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2000-03-13 | | Release date: | 2000-12-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Monomeric sarcosine oxidase: 1. Flavin reactivity and active site binding determinants.
Biochemistry, 39, 2000
|
|
3SJ8
 
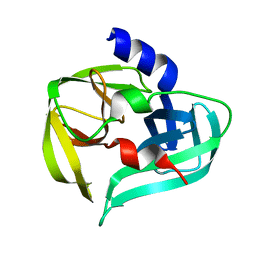 | | Crystal structure of the 3C protease from coxsackievirus A16 | | Descriptor: | 3C protease | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
3SJO
 
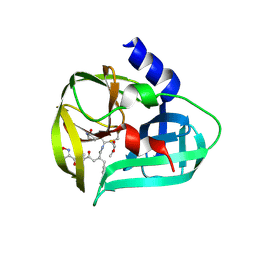 | | structure of EV71 3C in complex with Rupintrivir (AG7088) | | Descriptor: | 3C protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
3B5H
 
 | |
4FSJ
 
 | | Crystal structure of the virus like particle of Flock House virus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Speir, J.A, Chen, Z, Reddy, V.S, Johnson, J.E. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study of virus assembly intermediates reveals maturation event sequence and a staging position for externalized lytic peptides
to be published, 2012
|
|
3S6P
 
 | |
3SJI
 
 | | crystal structure of CVA16 3C in complex with Rupintrivir (AG7088) | | Descriptor: | 3C protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, SODIUM ION | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
1DIQ
 
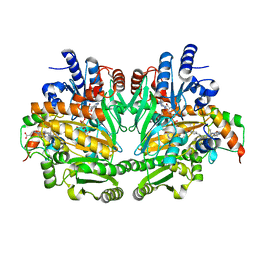 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE WITH SUBSTRATE BOUND | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.S, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1DJQ
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT C30A MUTANT OF TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
4GEE
 
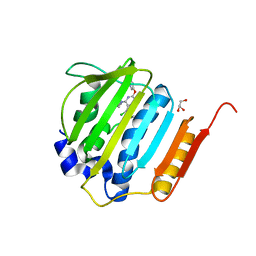 | | Pyrrolopyrimidine inhibitors of DNA gyrase B and topoisomerase IV, part I: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | (1R,5S,6s)-3-[5-chloro-6-ethyl-2-(pyrimidin-5-yloxy)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-3-azabicyclo[3.1.0]hexan-6-amine, DNA gyrase subunit B, GLYCEROL | | Authors: | Bensen, D.C, Chen, Z, Tari, L.W. | | Deposit date: | 2012-08-01 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3SJ9
 
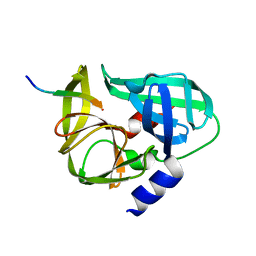 | | crystal structure of the C147A mutant 3C of CVA16 in complex with FAGLRQAVTQ peptide | | Descriptor: | 3C protease, FAGLRQAVTQ peptide | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
3SJK
 
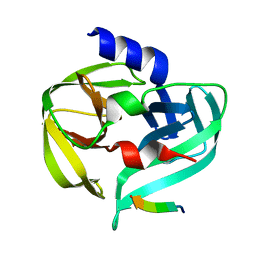 | | Crystal structure of the C147A mutant 3C from enterovirus 71 | | Descriptor: | 3C protease, KPVLRTATVQGPSLDF peptide | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
2HWL
 
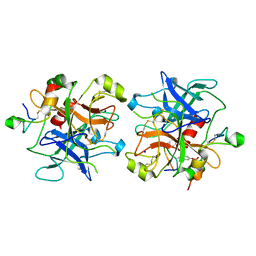 | | Crystal structure of thrombin in complex with fibrinogen gamma' peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen gamma' peptide, Prothrombin, ... | | Authors: | Pineda, A.O, Chen, Z.W, Marino, F, Mathews, F.S, Mosesson, M.W, Di Cera, E. | | Deposit date: | 2006-08-01 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thrombin in complex with fibrinogen gamma' peptide.
Biophys.Chem., 125, 2007
|
|
3SQH
 
 | |
3SQE
 
 | |
2LKK
 
 | | Human L-FABP in complex with oleate | | Descriptor: | Fatty acid-binding protein, liver, OLEIC ACID | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2011-10-13 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
2OV0
 
 | | Structure of the blue copper protein Amicyanin to 0.75 A resolution | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Carrell, C.J, Davidson, V.L, Chen, Z, Cunane, L.M, Trickey, P, Mathews, F.S. | | Deposit date: | 2007-02-12 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Ultrahigh resolution studies of amicyanin
TO BE PUBLISHED
|
|
3KUO
 
 | | X-ray structure of the metcyano form of dehaloperoxidase from amphitrite ornata: evidence for photoreductive lysis of iron-cyanide bond | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Serrano, V.S, Chen, Z, Gaff, J.F, Rose, R, Franzen, S. | | Deposit date: | 2009-11-27 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | X-ray structure of the metcyano form of dehaloperoxidase from Amphitrite ornata: evidence for photoreductive dissociation of the iron-cyanide bond.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5E7R
 
 | | Crystal structure of TL10-81 bound to TAK1-TAB1 | | Descriptor: | 2-chloro-N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}acetamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Gurbani, D, Hunter, J.C, Tan, L, Chen, Z, Westover, K.D. | | Deposit date: | 2015-10-13 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
1BFZ
 
 | | BOUND CONFORMATION OF N-TERMINAL CLEAVAGE PRODUCT PEPTIDE MIMIC (P1-P9 OF RELEASE SITE) WHILE BOUND TO HCMV PROTEASE AS DETERMINED BY TRANSFERRED NOESY EXPERIMENTS (P1-P5 SHOWN ONLY), NMR, 32 STRUCTURES | | Descriptor: | HCMV PROTEASE R-SITE N-TERMINAL CLEAVAGE PRODUCT | | Authors: | Laplante, S.R, Aubry, N, Bonneau, P.R, Cameron, D.R, Lagace, L, Massariol, M.-J, Montpetit, H, Ploufe, C, Kawai, S.H, Fulton, B.D, Chen, Z, Ni, F. | | Deposit date: | 1998-05-25 | | Release date: | 1999-05-25 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human cytomegalovirus protease complexes its substrate recognition sequences in an extended peptide conformation.
Biochemistry, 37, 1998
|
|
3EE0
 
 | | Crystal Structure of the W215A/E217A Mutant of Human Thrombin (space group P2(1)2(1)2(1)) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis for the Kinetic Differences of the W215A/E217A Mutant of Human and Murine Thrombin
To be Published
|
|
3KPP
 
 | | Crystal Structure of HLA B*4405 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, EEYLQAFTY, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KPN
 
 | | Crystal Structure of HLA B*4403 in complex with EEYLQAFTY a self peptide from the ABCD3 protein | | Descriptor: | Beta-2-microglobulin, EEYLQAFTY, self peptide from the ATP binding cassette protein ABCD3, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
