4PR3
 
 | | Crystal structure of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine nucleosidase / s-adenosylhomocysteine nucleosidase, ADENINE, GLYCEROL, ... | | Authors: | Zhang, X.C, Kang, X.S, Zhao, Y, Jiang, D.H, Li, X.M, Chen, Z.L. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal structure and biochemical studies of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Biochem.Biophys.Res.Commun., 446, 2014
|
|
6IRO
 
 | | the crosslinked complex of ISWI-nucleosome in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2018-11-13 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat. Struct. Mol. Biol., 26, 2019
|
|
5X4G
 
 | | Crystal structure of Fab fragment of anti-CD147 monoclonal antibody 6H8 | | Descriptor: | 6H8 Fab heavy chain, 6H8 Fab light chain | | Authors: | Lin, P, Zhang, M.-Y, Chen, X, Ye, S, Yu, X.-L, Zhang, R.-G, Zhu, P, Chen, Z.-N. | | Deposit date: | 2017-02-13 | | Release date: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CD147 C2 domain in complex with Fab of its monoclonal antibody
To Be Published
|
|
4H6S
 
 | | Crystal structure of thrombin mutant E14eA/D14lA/E18A/S195A | | Descriptor: | Prothrombin, SODIUM ION | | Authors: | Pozzi, N, Chen, Z, Zapata, F, Pelc, L.A, Di Cera, E. | | Deposit date: | 2012-09-19 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Autoactivation of thrombin precursors.
J.Biol.Chem., 288, 2013
|
|
1MG2
 
 | | MUTATION OF ALPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN | | Descriptor: | Amicyanin, COPPER (II) ION, CYTOCHROME C-L, ... | | Authors: | Sun, D, Chen, Z.W, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2002-08-14 | | Release date: | 2002-12-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | MUTATION OF AlPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN
Biochemistry, 41, 2002
|
|
2HWL
 
 | | Crystal structure of thrombin in complex with fibrinogen gamma' peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fibrinogen gamma' peptide, Prothrombin, ... | | Authors: | Pineda, A.O, Chen, Z.W, Marino, F, Mathews, F.S, Mosesson, M.W, Di Cera, E. | | Deposit date: | 2006-08-01 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of thrombin in complex with fibrinogen gamma' peptide.
Biophys.Chem., 125, 2007
|
|
1MG3
 
 | | MUTATION OF ALPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN | | Descriptor: | Amicyanin, COPPER (II) ION, CYTOCHROME C-L, ... | | Authors: | Sun, D, Chen, Z.W, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2002-08-14 | | Release date: | 2002-12-11 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | MUTATION OF ALPHA PHE55 OF METHYLAMINE DEHYDROGENASE ALTERS THE REORGANIZATION ENERGY AND ELECTRONIC COUPLING FOR ITS ELECTRON TRANSFER REACTION WITH AMICYANIN
Biochemistry, 41, 2002
|
|
2H47
 
 | | Crystal Structure of an Electron Transfer Complex Between Aromatic Amine Dephydrogenase and Azurin from Alcaligenes Faecalis (Form 1) | | Descriptor: | Aromatic Amine Dehydrogenase, Azurin, COPPER (II) ION | | Authors: | Sukumar, N, Chen, Z, Leys, D, Scrutton, N.S, Ferrati, D, Merli, A, Rossi, G.L, Bellamy, H.D, Chistoserdov, A, Davidson, V.L, Mathews, F.S. | | Deposit date: | 2006-05-23 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of an Electron Transfer Complex between Aromatic Amine Dehydrogenase and Azurin from Alcaligenes faecalis.
Biochemistry, 45, 2006
|
|
2L68
 
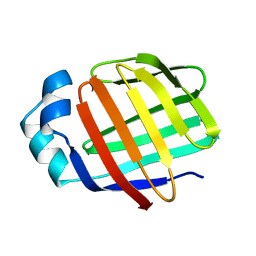 | | Solution Structure of Human Holo L-FABP | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
2L67
 
 | | Solution Structure of Human Apo L-FABP | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
4FWJ
 
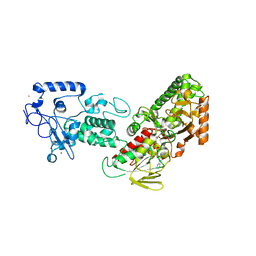 | | Native structure of LSD2/AOF1/KDM1b in spacegroup of I222 at 2.9A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, PHOSPHATE ION, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
5EE3
 
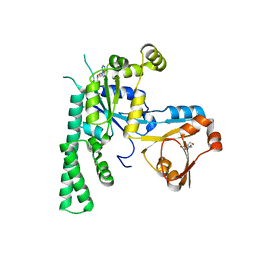 | | COMPLEX STRUCTURE OF OSYCHF1 WITH AMP-PNP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4FWE
 
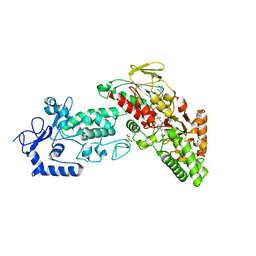 | | Native structure of LSD2 /AOF1/KDM1b in spacegroup of C2221 at 2.13A | | Descriptor: | CITRATE ANION, FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1B, ... | | Authors: | Zhang, Q, Chen, Z. | | Deposit date: | 2012-07-01 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b
Cell Res., 23, 2013
|
|
5EE0
 
 | | Crystal structure of OsYchF1 at pH 6.5 | | Descriptor: | Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2015-10-22 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ATP binding by the P-loop NTPase OsYchF1 (an unconventional G protein) contributes to biotic but not abiotic stress responses
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4M1L
 
 | | Complex of IQCG and Ca2+-bound CaM | | Descriptor: | CALCIUM ION, Calmodulin, IQ domain-containing protein G, ... | | Authors: | Liang, W.X, Chen, L.T, Chen, Z, Chen, S.J, Chen, S. | | Deposit date: | 2013-08-03 | | Release date: | 2014-05-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and molecular features of the calmodulin-interacting protein IQCG required for haematopoiesis in zebrafish
Nat Commun, 5, 2014
|
|
6AOG
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF, and 5-(4-chlorophenyl)-6-ethylpyrimidine-2,4-diamine (pyrimethamine) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, ... | | Authors: | Thomas, S.B, Li, Y, Chen, Z, Lu, H. | | Deposit date: | 2017-08-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification of selective, brain penetrant Toxoplasma gondii dihydrofolate reductase inhibitors for the treatment of toxoplasmosis
To Be Published
|
|
6IY3
 
 | | Structure of Snf2-MMTV-A nucleosome complex at shl-2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
6IY2
 
 | | Structure of Snf2-MMTV-A nucleosome complex at shl2 in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-12-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
6AOH
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF, and 5-(4-(3-(2-methoxypyrimidin-5-yl)phenyl)piperazin-1-yl)pyrimidine-2,4-diamine (TRC-2533) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-{4-[3-(2-methoxypyrimidin-5-yl)phenyl]piperazin-1-yl}pyrimidine-2,4-diamine, ... | | Authors: | Thomas, S.B, Li, Y, Chen, Z, Lu, H. | | Deposit date: | 2017-08-16 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Identification of selective, brain penetrant Toxoplasma gondii dihydrofolate reductase inhibitors for the treatment of toxoplasmosis
To Be Published
|
|
3R3G
 
 | | Structure of human thrombin with residues 145-150 of murine thrombin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Thrombin Heavy Chain, ... | | Authors: | Pozzi, N, Chen, R, Chen, Z, Bah, A, Di Cera, E. | | Deposit date: | 2011-03-15 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rigidification of the autolysis loop enhances Na(+) binding to thrombin.
Biophys.Chem., 159, 2011
|
|
1NWO
 
 | |
4MLF
 
 | | Crystal structure for the complex of thrombin mutant D102N and hirudin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Hirudin variant-1, ... | | Authors: | Vogt, A.D, Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Essential role of conformational selection in ligand binding.
Biophys.Chem., 186C, 2014
|
|
1JJU
 
 | | Structure of a Quinohemoprotein Amine Dehydrogenase with a Unique Redox Cofactor and Highly Unusual Crosslinking | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, QUINOHEMOPROTEIN AMINE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Datta, S, Mori, Y, Takagi, K, Kawaguchi, K, Chen, Z.-W, Kano, K, Ikeda, T, Okajima, T, Kuroda, S, Tanizawa, K, Mathews, F.S. | | Deposit date: | 2001-07-09 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5IRE
 
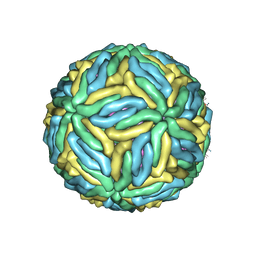 | | The cryo-EM structure of Zika Virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sirohi, D, Chen, Z, Sun, L, Klose, T, Pierson, T, Rossmann, M, Kuhn, R. | | Deposit date: | 2016-03-13 | | Release date: | 2016-03-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom resolution cryo-EM structure of Zika virus.
Science, 352, 2016
|
|
1L9C
 
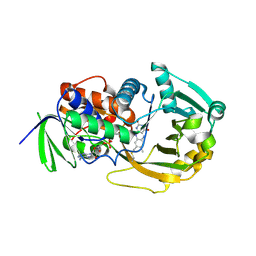 | | Role of Histidine 269 in Catalysis by Monomeric Sarcosine Oxidase | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric Sarcosine Oxidase, ... | | Authors: | Zhao, G, Song, H, Chen, Z.-w, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-08-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Monomeric sarcosine oxidase: role of histidine 269 in catalysis.
Biochemistry, 41, 2002
|
|
