3SQE
 
 | |
3CX6
 
 | |
1GG4
 
 | |
1T8P
 
 | | Crystal structure of Human erythrocyte 2,3-bisphosphoglycerate mutase | | Descriptor: | Bisphosphoglycerate mutase | | Authors: | Wang, Y, Wei, Z, Bian, Q, Cheng, Z, Wan, M, Liu, L, Gong, W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human bisphosphoglycerate mutase
J.Biol.Chem., 279, 2004
|
|
4TQ9
 
 | | Crystal Structure of a GDP-bound G12V Oncogenic Mutant of Human GTPase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
3S6P
 
 | |
3SJI
 
 | | crystal structure of CVA16 3C in complex with Rupintrivir (AG7088) | | Descriptor: | 3C protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER, SODIUM ION | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
4TQA
 
 | | Crystal Structure of a GDP-bound G13D Oncogenic Mutant of Human GTPase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
2PKA
 
 | | REFINED 2 ANGSTROMS X-RAY CRYSTAL STRUCTURE OF PORCINE PANCREATIC KALLIKREIN A, A SPECIFIC TRYPSIN-LIKE SERINE PROTEINASE. CRYSTALLIZATION, STRUCTURE DETERMINATION, CRYSTALLOGRAPHIC REFINEMENT, STRUCTURE AND ITS COMPARISON WITH BOVINE TRYPSIN | | Descriptor: | BENZAMIDINE, KALLIKREIN A | | Authors: | Bode, W, Chen, Z. | | Deposit date: | 1984-05-21 | | Release date: | 1984-07-19 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refined 2 A X-ray crystal structure of porcine pancreatic kallikrein A, a specific trypsin-like serine proteinase. Crystallization, structure determination, crystallographic refinement, structure and its comparison with bovine trypsin.
J.Mol.Biol., 164, 1983
|
|
3GIC
 
 | | Structure of thrombin mutant delta(146-149e) in the free form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin heavy chain, Thrombin light chain | | Authors: | Bah, A, Carrell, C.J, Chen, Z, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-03-05 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Stabilization of the E* form turns thrombin into an anticoagulant.
J.Biol.Chem., 284, 2009
|
|
2LKK
 
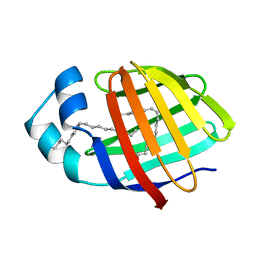 | | Human L-FABP in complex with oleate | | Descriptor: | Fatty acid-binding protein, liver, OLEIC ACID | | Authors: | Cai, J, Luecke, C, Chen, Z, Qiao, Y, Klimtchuk, E.S, Hamilton, J.A. | | Deposit date: | 2011-10-13 | | Release date: | 2012-06-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of human liver fatty acid binding protein: fatty acid binding revisited.
Biophys.J., 102, 2012
|
|
5E7R
 
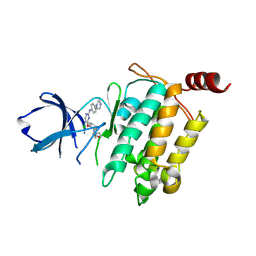 | | Crystal structure of TL10-81 bound to TAK1-TAB1 | | Descriptor: | 2-chloro-N-{2-[(5-chloro-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)oxy]phenyl}acetamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Gurbani, D, Hunter, J.C, Tan, L, Chen, Z, Westover, K.D. | | Deposit date: | 2015-10-13 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-guided development of covalent TAK1 inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
2MLD
 
 | |
5CL1
 
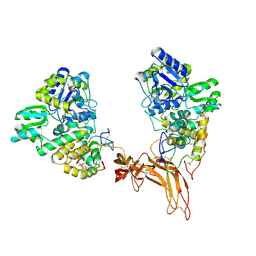 | | Complex structure of Norrin with human Frizzled 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Maltose-binding periplasmic protein,Norrin | | Authors: | Wang, Z, Ke, J, Shen, G, Cheng, Z, Xu, H.E, Xu, W. | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of the Norrin-Frizzled 4 interaction.
Cell Res., 25, 2015
|
|
2OV0
 
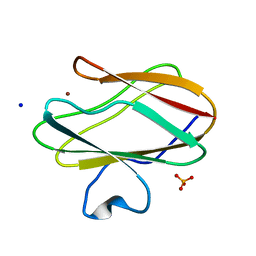 | | Structure of the blue copper protein Amicyanin to 0.75 A resolution | | Descriptor: | Amicyanin, COPPER (II) ION, PHOSPHATE ION, ... | | Authors: | Carrell, C.J, Davidson, V.L, Chen, Z, Cunane, L.M, Trickey, P, Mathews, F.S. | | Deposit date: | 2007-02-12 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Ultrahigh resolution studies of amicyanin
TO BE PUBLISHED
|
|
7V27
 
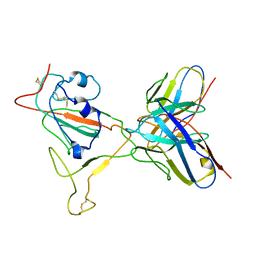 | | RBD/XG005 local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, XG005-VH, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | RBD/XG005 local refinement
To Be Published
|
|
5V8F
 
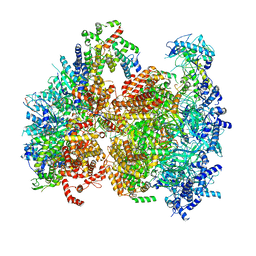 | | Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1 | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (39-MER), ... | | Authors: | Yuan, Z, Riera, A, Bai, L, Sun, J, Spanos, C, Chen, Z.A, Barbon, M, Rappsilber, J, Stillman, B, Speck, C, Li, H. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-10 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2P5B
 
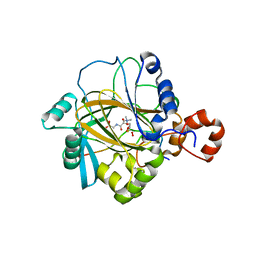 | | The complex structure of JMJD2A and trimethylated H3K36 peptide | | Descriptor: | FE (II) ION, Histone H3, JmjC domain-containing histone demethylation protein 3A, ... | | Authors: | Zhang, G, Chen, Z, Zang, J, Hong, X, Shi, Y. | | Deposit date: | 2007-03-14 | | Release date: | 2007-06-12 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of the recognition of a methylated histone tail by JMJD2A.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2PMV
 
 | | Crystal Structure of Human Intrinsic Factor- Cobalamin Complex at 2.6 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COBALAMIN, Gastric intrinsic factor | | Authors: | Mathews, F.S, Gordon, M.M, Chen, Z, Rajashankar, K.R, Ealick, S.E, Alpers, D.H, Sukumar, N. | | Deposit date: | 2007-04-23 | | Release date: | 2007-10-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human intrinsic factor: Cobalamin complex at 2.6-A resolution
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
3KPQ
 
 | | Crystal Structure of HLA B*4405 in complex with EEYLKAWTF, a mimotope | | Descriptor: | Beta-2-microglobulin, EEYLKAWTF, mimotope peptide, ... | | Authors: | Macdonald, W.A, Chen, Z, Gras, S, Archbold, J.K, Tynan, F.E, Clements, C.S, Bharadwaj, M, Kjer-Nielsen, L, Saunders, P.M, Wilce, M.C, Crawford, F, Stadinsky, B, Jackson, D, Brooks, A.G, Purcell, A.W, Kappler, J.W, Burrows, S.R, Rossjohn, J, McCluskey, J. | | Deposit date: | 2009-11-16 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | T cell allorecognition via molecular mimicry.
Immunity, 31, 2009
|
|
3KUN
 
 | | X-ray structure of the metcyano form of dehaloperoxidase from amphitrite ornata: evidence for photoreductive lysis of iron-cyanide bond | | Descriptor: | CYANIDE ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Serrano, V.S, Chen, Z, Gaff, J.F, Rose, R, Franzen, S. | | Deposit date: | 2009-11-27 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | X-ray structure of the metcyano form of dehaloperoxidase from Amphitrite ornata: evidence for photoreductive dissociation of the iron-cyanide bond.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2QFK
 
 | | X-ray Crystal Structure Analysis of the Binding Site in the Ferric and Oxyferrous Forms of the Recombinant Heme Dehaloperoxidase Cloned from Amphitrite ornata | | Descriptor: | AMMONIUM ION, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | de Serrano, V.S, Chen, Z, Davis, M.F, Franzen, S. | | Deposit date: | 2007-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray crystal structural analysis of the binding site in the ferric and
oxyferrous forms of the recombinant heme dehaloperoxidase cloned from Amphitrite ornata
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2MLA
 
 | |
2R9E
 
 | |
5TO3
 
 | | Crystal structure of thrombin mutant W215A/E217A fused to EGF456 of thrombomodulin via a 31-residue linker and bound to PPACK | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Barranco-Medina, S, Murphy, M, Pelc, L, Chen, Z, Di Cera, E, Pozzi, N. | | Deposit date: | 2016-10-16 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Rational Design of Protein C Activators.
Sci Rep, 7, 2017
|
|
