4WA7
 
 | | Crystal Structure of a GDP-bound Q61L Oncogenic Mutant of Human GT- Pase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Manandhar, A, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2014-08-28 | | Release date: | 2015-06-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations.
Mol Cancer Res., 13, 2015
|
|
4DSB
 
 | | Complex Structure of Abscisic Acid Receptor PYL3 with (+)-ABA in Spacegroup of I 212121 at 2.70A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3 | | Authors: | Zhang, X, Zhang, Q, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
7XA3
 
 | | Cryo-EM structure of the CCL2 bound CCR2-Gi complex | | Descriptor: | C-C motif chemokine 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
7X9Y
 
 | | Cryo-EM structure of the apo CCR3-Gi complex | | Descriptor: | C-C chemokine receptor type 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
1SGI
 
 | | Crystal structure of the anticoagulant slow form of thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, thrombin | | Authors: | Pineda, A.O, Carrell, C.J, Bush, L.A, Prasad, S, Caccia, S, Chen, Z.W, Mathews, F.S, Di Cera, E. | | Deposit date: | 2004-02-23 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular dissection of na+ binding to thrombin.
J.Biol.Chem., 279, 2004
|
|
1INP
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | INOSITOL POLYPHOSPHATE 1-PHOSPHATASE, MAGNESIUM ION | | Authors: | York, J.D, Ponder, J.W, Chen, Z, Mathews, F.S, Majerus, P.W. | | Deposit date: | 1994-10-04 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of inositol polyphosphate 1-phosphatase at 2.3-A resolution.
Biochemistry, 33, 1994
|
|
1Z71
 
 | | thrombin and P2 pyridine N-oxide inhibitor complex structure | | Descriptor: | Hirudin IIIB', L17, thrombin | | Authors: | Nantermet, P.G, Burgey, C.S, Robinson, K.A, Pellicore, J.M, Newton, C.L, Deng, J.Z, Lyle, T.A, Selnick, H.G, Lewis, S.D, Lucas, B.J, Krueger, J.A, Miller-Stein, C, White, R.B, Wong, B, McMasters, D.R, Wallace, A.A, Lynch Jr, J.J, Yan, Y, Chen, Z, Kuo, L, Gardell, S.J, Shafer, J.A, Vacca, J.P. | | Deposit date: | 2005-03-23 | | Release date: | 2005-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | P(2) pyridine N-oxide thrombin inhibitors: a novel peptidomimetic scaffold
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2010-08-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
4DSC
 
 | | Complex structure of abscisic acid receptor PYL3 with (+)-ABA in spacegroup of H32 at 1.95A | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION | | Authors: | Zhang, X, Chen, Z. | | Deposit date: | 2012-02-18 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
7UQ2
 
 | | Vs.4 from T4 phage in complex with cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, CALCIUM ION, Vs.4 | | Authors: | Jenson, J.M, Chen, Z.J. | | Deposit date: | 2022-04-18 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ubiquitin-like conjugation by bacterial cGAS enhances anti-phage defence.
Nature, 616, 2023
|
|
6MDW
 
 | | Mechanism of protease dependent DPC repair | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CITRATE ANION, ... | | Authors: | Li, F, Raczynska, J, Chen, Z, Yu, H. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insight into DNA-Dependent Activation of Human Metalloprotease Spartan.
Cell Rep, 26, 2019
|
|
7Y9I
 
 | | Complex structure of AtYchF1 with ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, MAGNESIUM ION, Obg-like ATPase 1 | | Authors: | Li, X, Chen, Z. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Co-crystalization reveals the interaction between AtYchF1 and ppGpp.
Front Mol Biosci, 9, 2022
|
|
8JX7
 
 | | Cryo-EM structure of human ABC transporter ABCC2 | | Descriptor: | ATP-binding cassette sub-family C member 2 | | Authors: | Mao, Y.X, Chen, Z.P, Wang, L, Hou, W.T, Chen, Y.X, Zhou, C.Z. | | Deposit date: | 2023-06-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Transport mechanism of human bilirubin transporter ABCC2 tuned by the inter-module regulatory domain.
Nat Commun, 15, 2024
|
|
8Y6V
 
 | | Near-atomic structure of icosahedrally averaged jumbo bacteriophage PhiKZ capsid | | Descriptor: | gp119, gp120, gp162, ... | | Authors: | Yang, Y, Shao, Q, Guo, M, Han, L, Zhao, X, Wang, A, Li, X, Wang, B, Pan, J, Chen, Z, Fokine, A, Sun, L, Fang, Q. | | Deposit date: | 2024-02-03 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capsid structure of bacteriophage Phi KZ provides insights into assembly and stabilization of jumbo phages.
Nat Commun, 15, 2024
|
|
7VJV
 
 | | Human AlkB homolog ALKBH6 in complex with alpha-katoglutarate and Mn | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, MANGANESE (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7VJS
 
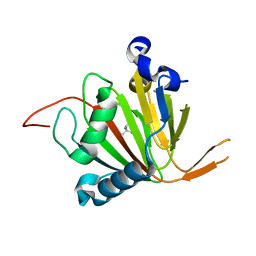 | | Human AlkB homolog ALKBH6 in complex with Tris and Ni | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-ketoglutarate-dependent dioxygenase alkB homolog 6, NICKEL (II) ION | | Authors: | Ma, L, Chen, Z. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Structural insights into the interactions and epigenetic functions of human nucleic acid repair protein ALKBH6.
J.Biol.Chem., 298, 2022
|
|
7WK1
 
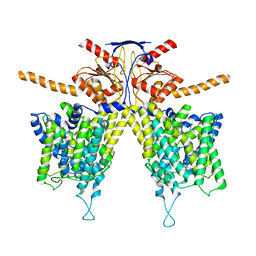 | | Mouse Pendrin bound chloride in inward state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WL7
 
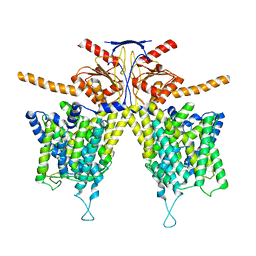 | |
7WL2
 
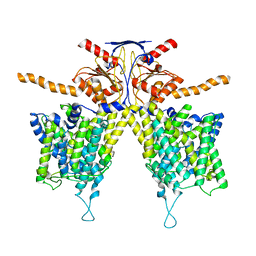 | |
7WLE
 
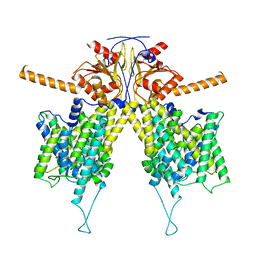 | |
7WL9
 
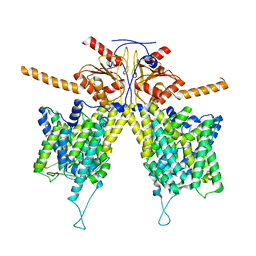 | | Mouse Pendrin in chloride and bicarbonate in asymmetric state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-12 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WLA
 
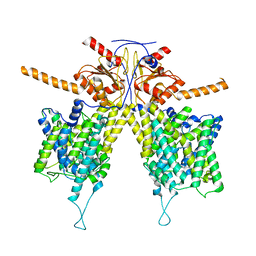 | |
7WK7
 
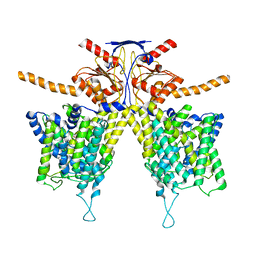 | | Mouse Pendrin bound bicarbonate in inward state | | Descriptor: | BICARBONATE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-05-17 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7CXZ
 
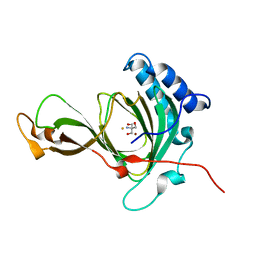 | | crystal structure of pco2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, NICKEL (II) ION, ... | | Authors: | Guo, Q, Xu, C, Liao, S, Chen, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Molecular basis for cysteine oxidation by plant cysteine oxidases from Arabidopsis thaliana.
J.Struct.Biol., 213, 2021
|
|
7EV8
 
 | |
