6S0O
 
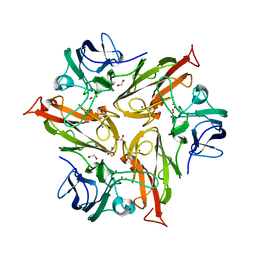 | | Crystal Structure of Two-Domain Laccase from Streptomyces griseoflavus produced at 0.25 mM copper sulfate in growth medium | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2019-06-17 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Investigations of Accessibility of T2/T3 Copper Center of Two-Domain Laccase fromStreptomyces griseoflavusAc-993.
Int J Mol Sci, 20, 2019
|
|
7BHS
 
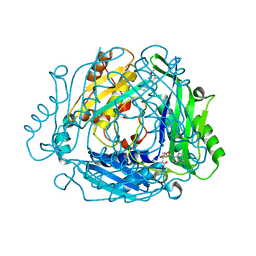 | | Crystal structure of MAT2a with quinazoline fragment 2 bound in the allosteric site | | Descriptor: | 6-chloranyl-2-methoxy-4-phenyl-quinazoline, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
5XS4
 
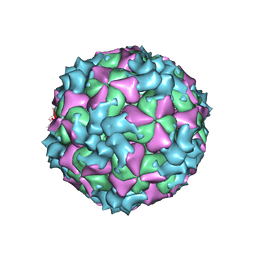 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5DY2
 
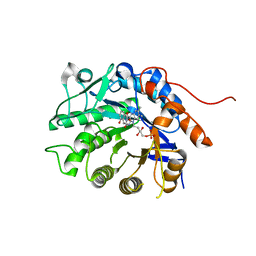 | | Crystal structure of Dbr2 with mutation M27L | | Descriptor: | 1,6-DIHYDROXY NAPHTHALENE, Artemisinic aldehyde Delta(11(13)) reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, L, Chen, T.T, Xu, Y.C. | | Deposit date: | 2015-09-24 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Crystal structure of Dbr2
to be published
|
|
3OUV
 
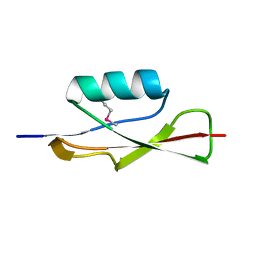 | |
5CWW
 
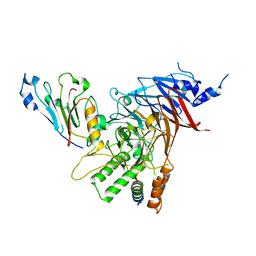 | | Crystal structure of the Chaetomium thermophilum heterotrimeric Nup82 NTD-Nup159 TAIL-Nup145N APD complex | | Descriptor: | Nucleoporin NUP145N, Nucleoporin NUP159, Nucleoporin NUP82 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
5CWV
 
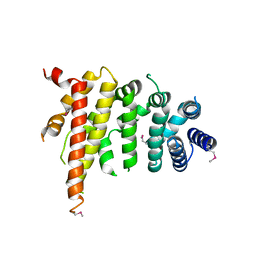 | | Crystal structure of Chaetomium thermophilum Nup192 TAIL domain | | Descriptor: | Nucleoporin NUP192 | | Authors: | Stuwe, T, Bley, C.J, Thierbach, K, Petrovic, S, Schilbach, S, Mayo, D.J, Perriches, T, Rundlet, E.J, Jeon, Y.E, Collins, L.N, Lin, D.H, Paduch, M, Koide, A, Lu, V, Fischer, J, Hurt, E, Koide, S, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.155 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
3VMQ
 
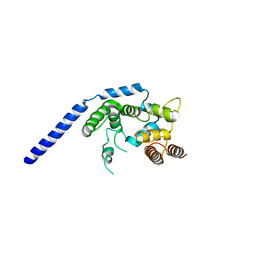 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase: Apoenzyme | | Descriptor: | MAGNESIUM ION, Monofunctional glycosyltransferase | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VMT
 
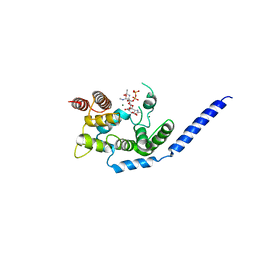 | | Crystal structure of Staphylococcus aureus membrane-bound transglycosylase in complex with a Lipid II analog | | Descriptor: | MAGNESIUM ION, Monofunctional glycosyltransferase, [(2R,3R,4R,5S,6R)-4-[(2R)-1-[[(2S)-1-[2-[2-[2-[5-[(3aS,4S,6aR)-2-oxidanylidene-1,3,3a,4,6,6a-hexahydrothieno[3,4-d]imidazol-4-yl]pentanoylamino]ethoxy]ethoxy]ethylamino]-1-oxidanylidene-propan-2-yl]amino]-1-oxidanylidene-propan-2-yl]oxy-3-acetamido-5-[(2S,3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-(hydroxymethyl)oxan-2-yl] [oxidanyl(3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,14,18,22,26,30,34,38,42-undecaenoxy)phosphoryl] hydrogen phosphate | | Authors: | Huang, C.Y, Shih, H.W, Lin, L.Y, Tien, Y.W, Cheng, T.J.R, Cheng, W.C, Wong, C.H, Ma, C. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Crystal structure of Staphylococcus aureus transglycosylase in complex with a lipid II analog and elucidation of peptidoglycan synthesis mechanism
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1XZ0
 
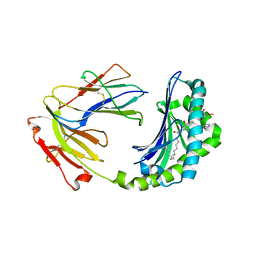 | | Crystal structure of CD1a in complex with a synthetic mycobactin lipopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(HYDROXY-HEXADECANOYL-AMINO)-2-{[(4S)-2-(2-HYDROXY-PHENYL)-4,5-DIHYDRO-OXAZOLE-4-CARBONYL]-AMINO}-HEXANOIC ACID 2-[(3S)-1-(TERT-BUTYL-DIPHENYL-SILANYLOXY)-2-OXO-AZEPAN-3-YLCARBAMOYL]-(1S)-1-METHYL-ETHYL ESTER, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Crispin, M.D, Bowden, T.A, Young, D.C, Cheng, T.Y, Hu, J, Costello, C.E, Miller, M.J, Moody, D.B, Wilson, I.A. | | Deposit date: | 2004-11-11 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism of Lipopeptide Presentation by CD1a.
Immunity, 22, 2005
|
|
4ZRA
 
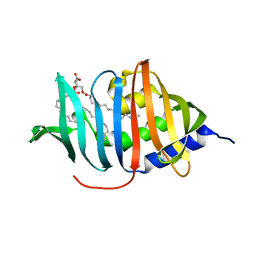 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS LPRG BINDING TO TRIACYLGLYCERIDE | | Descriptor: | Lipoprotein LprG, Tripalmitoylglycerol | | Authors: | Martinot, A.J, Farrow, M, Bai, L, Layre, E, Cheng, T.Y, Tsai, J.H.C, Iqbal, J, Annand, J, Sullivan, Z, Hussain, M, Sacchettini, J, Moody, D.B, Seeliger, J, Rubin, E.J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-05-12 | | Release date: | 2016-02-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis.
Plos Pathog., 12, 2016
|
|
7Z2X
 
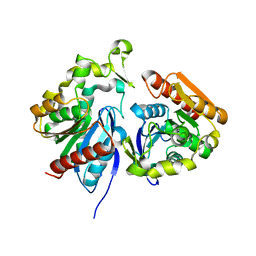 | | Wild-type ferulic acid esterase from Lactobacillus buchneri | | Descriptor: | Ferulic acid esterase | | Authors: | Mogodiniyai, K.K, Reichenbach, T, Kalyani, D.C, Keskitalo, M.M, Divne, C. | | Deposit date: | 2022-03-01 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the feruloyl esterase from Lentilactobacillus buchneri reveals a novel homodimeric state.
Front Microbiol, 13, 2022
|
|
7Z2U
 
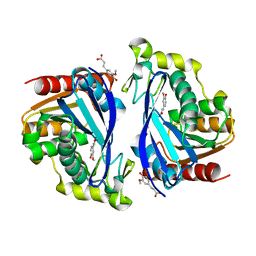 | | Wild-type ferulic acid esterase from Lactobacillus buchneri in complex with ferulate | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, Ferulic acid esterase | | Authors: | Mogodiniyai, K.K, Reichenbach, T, Kalyani, D.C, Keskitalo, M.M, Divne, C. | | Deposit date: | 2022-02-28 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the feruloyl esterase from Lentilactobacillus buchneri reveals a novel homodimeric state.
Front Microbiol, 13, 2022
|
|
7Z2V
 
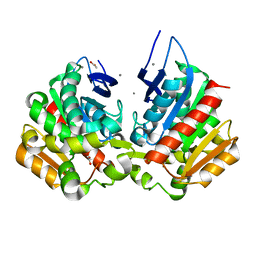 | | Ferulic acid esterase variant S114A from Lactobacillus buchneri | | Descriptor: | ACETATE ION, CALCIUM ION, Ferulic acid esterase | | Authors: | Mogodiniyai, K.K, Reichenbach, T, Kalyani, D.C, Keskitalo, M.M, Divne, C. | | Deposit date: | 2022-02-28 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of the feruloyl esterase from Lentilactobacillus buchneri reveals a novel homodimeric state.
Front Microbiol, 13, 2022
|
|
7V3R
 
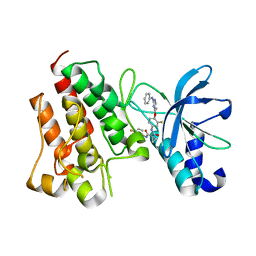 | | Crystal structure of CMET in complex with a novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, ~{N}1'-[3-fluoranyl-4-(2-phenylazanylpyrimidin-4-yl)oxy-phenyl]-~{N}1-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Su, H.X, Liu, Q.F, Chen, T.T, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 10H-Benzo[b]pyrido[2,3-e][1,4]oxazine AXL Inhibitors via Structure-Based Drug Design Targeting c-Met Kinase
J.Med.Chem., 66, 2023
|
|
7V3S
 
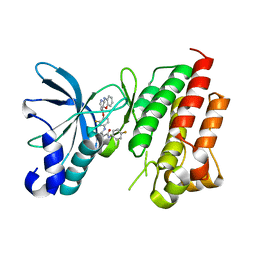 | | Crystal structure of CMET in complex with a novel inhibitor | | Descriptor: | Hepatocyte growth factor receptor, ~{N}1'-[3-fluoranyl-4-(10~{H}-pyrido[3,2-b][1,4]benzoxazin-4-yloxy)phenyl]-~{N}1-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Su, H.X, Liu, Q.F, Chen, T.T, Li, M.J, Xu, Y.C. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of 10H-Benzo[b]pyrido[2,3-e][1,4]oxazine AXL Inhibitors via Structure-Based Drug Design Targeting c-Met Kinase
J.Med.Chem., 66, 2023
|
|
3JAU
 
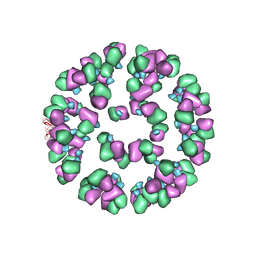 | | The cryoEM map of EV71 mature viron in complex with the Fab fragment of antibody D5 | | Descriptor: | Capsid protein VP1, Heavy chain of Fab fragment variable region of antibody D5, Light chain of Fab fragment variable region of antibody D5 | | Authors: | Fan, C, Ye, X.H, Ku, Z.Q, Zuo, T, Kong, L.L, Zhang, C, Shi, J.P, Liu, Q.W, Chen, T, Zhang, Y.Y, Jiang, W, Zhang, L.Q, Huang, Z, Cong, Y. | | Deposit date: | 2015-06-24 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural Basis for Recognition of Human Enterovirus 71 by a Bivalent Broadly Neutralizing Monoclonal Antibody
Plos Pathog., 12, 2016
|
|
8AF1
 
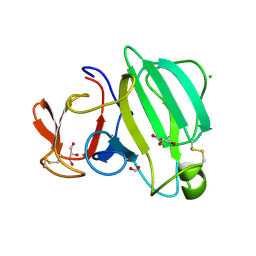 | | Beta-Lytic Protease from Lysobacter capsici | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kudryakova, I.V, Afoshin, A.S, Vasilyeva, N.V. | | Deposit date: | 2022-07-15 | | Release date: | 2023-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural and Functional Characterization of beta-lytic Protease from Lysobacter capsici VKM B-2533 T.
Int J Mol Sci, 23, 2022
|
|
5JD5
 
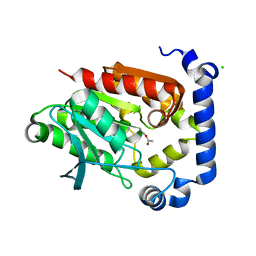 | | Crystal structure of MGS-MilE3, an alpha/beta hydrolase enzyme from the metagenome of pyrene-phenanthrene enrichment culture with sediment sample of Milazzo Harbor, Italy | | Descriptor: | CHLORIDE ION, MGS-MilE3 | | Authors: | Stogios, P.J, Xu, X, Cui, H, Martinez-Martinez, M, Chernikova, T.N, Golyshin, P.N, Yakimov, M.M, Ferrer, M, Savchenko, A. | | Deposit date: | 2016-04-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of MGS-MilE3, an alpha/beta hydrolase enzyme from the metagenome of pyrene-phenanthrene enrichment culture with sediment sample of Milazzo Harbor, Italy
To Be Published
|
|
5Y6K
 
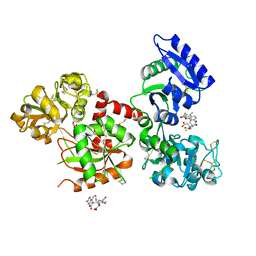 | | Human serum trnasferrin bound to a fluorescent probe | | Descriptor: | (2S)-6-[2-(7-azido-4-methyl-2-oxidanylidene-chromen-3-yl)ethanoylamino]-2-[bis(2-hydroxy-2-oxoethyl)amino]hexanoic acid, FE (III) ION, MALONATE ION, ... | | Authors: | Jiang, N, Cheng, T, Wang, M, Chan, G.C.F, Jin, L, Li, H, Sun, H. | | Deposit date: | 2017-08-12 | | Release date: | 2018-01-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Tracking iron-associated proteomes in pathogens by a fluorescence approach.
Metallomics, 10, 2018
|
|
2OHW
 
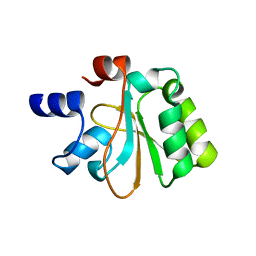 | | Crystal structure of the YueI protein from Bacillus subtilis | | Descriptor: | YueI protein | | Authors: | Bonanno, J.B, Jin, X, Mu, H, Dickey, M, Bain, K.T, Wu, B, Chen, T, Reyes, C, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-10 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the YueI protein from Bacillus subtilis
To be Published
|
|
6CT0
 
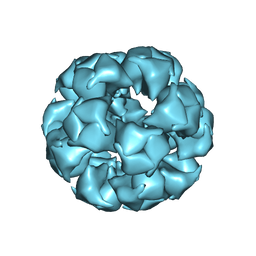 | | Atomic Structure of the E2 Inner Core of Human Pyruvate Dehydrogenase Complex | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Jiang, J, Baiesc, F.L, Hiromasa, Y, Yu, X, Hui, W.H, Dai, X, Roche, T.E, Zhou, Z.H. | | Deposit date: | 2018-03-21 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic Structure of the E2 Inner Core of Human Pyruvate Dehydrogenase Complex.
Biochemistry, 57, 2018
|
|
6TTB
 
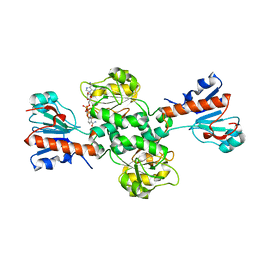 | | Crystal structure of NAD-dependent formate dehydrogenase from Staphylococcus aureus in complex with NAD | | Descriptor: | Formate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Boyko, K.M, Pometun, A.A, Nikolaeva, A.Y, Kargov, I.S, Yurchenko, T.S, Savin, S.S, Popov, V.O, Tishkov, V.I. | | Deposit date: | 2019-12-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of NAD-dependent formate dehydrogenase from Staphylococcus aureus in complex with NAD
To Be Published
|
|
1XR9
 
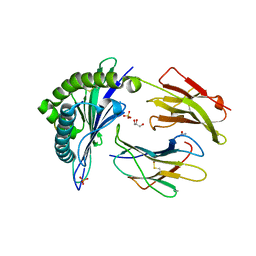 | | Crystal Structures of HLA-B*1501 in Complex with Peptides from Human UbcH6 and Epstein-Barr Virus EBNA-3 | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G, Blicher, T, Johannessen, B.R, Kristensen, O, Buus, S, Gajhede, M. | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Crystal structures of two peptide-HLA-B*1501 complexes; structural characterization of the HLA-B62 supertype
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3ZRG
 
 | | Crystal structure of RxLR effector PexRD2 from Phytophthora infestans | | Descriptor: | BROMIDE ION, PEXRD2 FAMILY SECRETED RXLR EFFECTOR PEPTIDE, PUTATIVE | | Authors: | King, S.R.F, Boutemy, L.S, Win, J, Hughes, R.K, Clarke, T.A, Blumenschein, T.M.A, Kamoun, S, Banfield, M.J. | | Deposit date: | 2011-06-16 | | Release date: | 2011-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Phytophthora Rxlr Effector Proteins: A Conserved But Adaptable Fold Underpins Functional Diversity.
J.Biol.Chem., 286, 2011
|
|
