5JEE
 
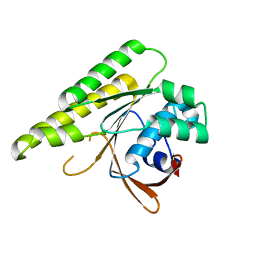 | | Apo-structure of humanised RadA-mutant humRadA26F | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JFG
 
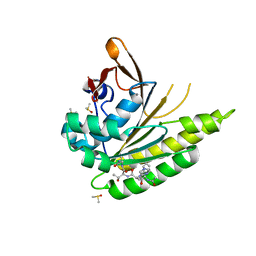 | | Structure of humanised RadA-mutant humRadA22F in complex with peptide FHTA | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Humanisation of RadA from Pyrococcus furiosus
To Be Published
|
|
4DVF
 
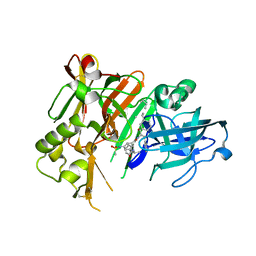 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S)-2,13-DIBENZYL-12-HYDROXY-3,5-DIMETHYL-8-(2-METHYLPROPYL)-15-(3-[(METHYLSULFONYL)AMINO]-5-{[(1R)-1-PHENYLETHYL]CARBAMOYL}PHENYL)-4,7,10,15-TETRAOXO-3,6,9,14-TETRAAZAPENTADECAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
4DV9
 
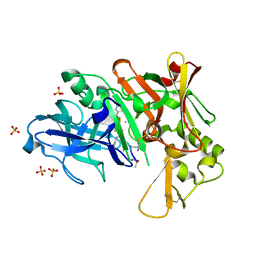 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2S)-1-[(2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-AMINO-3-OXOPROPYL)-2,13-DIBENZYL-12,22-DIHYDROXY-3,5,17-TRIMETHYL-8-(2-METHYLPROPYL)-4,7,10,15,18,21-HEXAOXO-19-(PROPAN-2-YL)-3,6,9,14,17,20-HEXAAZATRICOSAN-1-OYL]PYRROLIDINE-2-CARBOXYLATE (NON-PREFERRED NAME), SULFATE ION | | Authors: | Xu, Y.C, Chen, W.Y, Li, L, Chen, T.T. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
3TPP
 
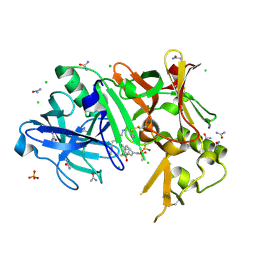 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE, ... | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TPJ
 
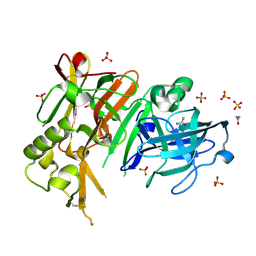 | | APO structure of BACE1 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, SULFATE ION, ... | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TPR
 
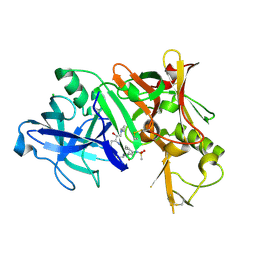 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3TPL
 
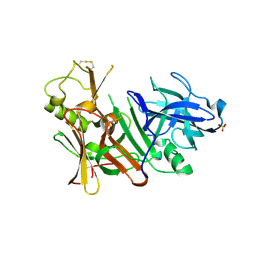 | | APO Structure of BACE1 | | Descriptor: | Beta-secretase 1, CHLORIDE ION, SULFATE ION | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
5KMD
 
 | | Structure of CavAb in complex with amlodipine | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-26 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
5KLB
 
 | | Crystal structure of the CavAb voltage-gated calcium channel(wild-type, 2.7A) | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CALCIUM ION, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-23 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
5KLS
 
 | | Structure of CavAb in complex with Br-dihydropyridine derivative UK-59811 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-25 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
5KMH
 
 | | Structure of CavAb in complex with Br-verapamil | | Descriptor: | (2~{R})-2-(2-bromophenyl)-5-[2-(3,4-dimethoxyphenyl)ethyl-methyl-amino]-2-propan-2-yl-pentanenitrile, 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIPALMITOYL-SN-GLYCERO-3-PHOSPHATE, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
5KMF
 
 | | Structure of CavAb in complex with nimodipine | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-26 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
5KLG
 
 | | Structure of CavAb(W195Y) in complex with Br-dihydropyridine derivative UK-59811 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein, ... | | Authors: | Tang, L, Gamal EL-Din, T.M, Swanson, T.M, Pryde, D.C, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs.
Nature, 537, 2016
|
|
2MRP
 
 | |
6ZB9
 
 | | Exo-beta-1,3-glucanase from moose rumen microbiome, wild type | | Descriptor: | Exo-beta-1,3-glucanase | | Authors: | Kalyani, D.C, Reichenbach, T, Aspeborg, H, Divne, C. | | Deposit date: | 2020-06-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A homodimeric bacterial exo-beta-1,3-glucanase derived from moose rumen microbiome shows a structural framework similar to yeast exo-beta-1,3-glucanases.
Enzyme.Microb.Technol., 143, 2021
|
|
6ZB8
 
 | | Exo-beta-1,3-glucanase from moose rumen microbiome, active site mutant E167Q/E295Q | | Descriptor: | Exo-beta-1,3-glucanase variant E167Q/E295Q, POLYETHYLENE GLYCOL (N=34) | | Authors: | Kalyani, D.C, Reichenbach, T, Aspeborg, H, Divne, C. | | Deposit date: | 2020-06-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A homodimeric bacterial exo-beta-1,3-glucanase derived from moose rumen microbiome shows a structural framework similar to yeast exo-beta-1,3-glucanases.
Enzyme.Microb.Technol., 143, 2021
|
|
8S8W
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8S8X
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Toyocamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
6LHC
 
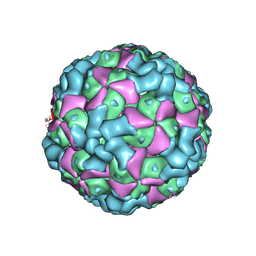 | | The cryo-EM structure of coxsackievirus A16 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHL
 
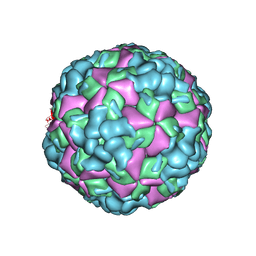 | | The cryo-EM structure of coxsackievirus A16 A-particle in complex with Fab 18A7 | | Descriptor: | VP1 protein, VP2 protein, VP3 protein | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-09 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6LHB
 
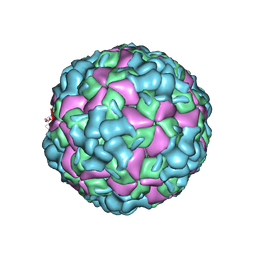 | | The cryo-EM structure of coxsackievirus A16 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6SYF
 
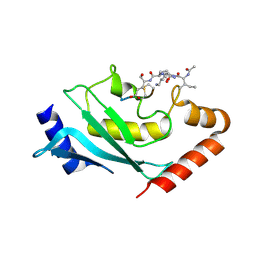 | | Human Ubc9 with covalent isopeptide ligand | | Descriptor: | ACE-ILE-LYS-GLN-GLU, ACE-LEU-ARG-LEU-ARG-GLY-CYS, SUMO-conjugating enzyme UBC9 | | Authors: | Hofmann, R, Akimoto, G, Wucherpfennig, T.G, Zeymer, C, Bode, J.W. | | Deposit date: | 2019-09-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Lysine acylation using conjugating enzymes for site-specific modification and ubiquitination of recombinant proteins.
Nat.Chem., 12, 2020
|
|
6ZIP
 
 | | Crystal Structure of Two-Domain Laccase mutant R240A from Streptomyces griseoflavus | | Descriptor: | COPPER (II) ION, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of positive charged residue in the proton-transfer mechanism of two-domain laccase from Streptomyces griseoflavus Ac-993.
J.Biomol.Struct.Dyn., 40, 2022
|
|
6ZIJ
 
 | | Crystal Structure of Two-Domain Laccase mutant R240H from Streptomyces griseoflavus | | Descriptor: | 1,2-ETHANEDIOL, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Kolyadenko, I.A. | | Deposit date: | 2020-06-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The role of positive charged residue in the proton-transfer mechanism of two-domain laccase from Streptomyces griseoflavus Ac-993.
J.Biomol.Struct.Dyn., 40, 2022
|
|
