4FGX
 
 | | Crystal structure of bace1 with novel inhibitor | | Descriptor: | Beta-secretase 1, DI(HYDROXYETHYL)ETHER, Inhibitor (2R,5S,8S,12S,13S,16S,19S,22S)-16-(3-amino-3-oxopropyl)-2,13-dibenzyl-12,22-dihydroxy-8-isobutyl-19-isopropyl-3,5,17-trimethyl-4,7,10,15,18,21-hexaoxo-3,6,9,14,17,20-hexaazatricosan-1-oic acid, ... | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2012-06-05 | | Release date: | 2013-01-16 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
3UQP
 
 | | Crystal structure of Bace1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2R)-1-[(6S,9S,12S,13S,17S,20S,23R)-9-(3-AMINO-3-OXOPROPYL)-12,23-DIBENZYL-13-HYDROXY-2,2,8,20,22-PENTAMETHYL-17-(2-METHYLPROPYL)-4,7,10,15,18,21,24-HEPTAOXO-6-(PROPAN-2-YL)-3-OXA-5,8,11,16,19,22-HEXAAZATETRACOSAN-24-YL]PYRROLIDINE-2-CARBOXYLATE, SULFATE ION | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
4FCO
 
 | | Crystal structure of bace1 with its inhibitor | | Descriptor: | Beta-secretase 1, N-[(2S,3R)-4-{[2-(1-benzylpiperidin-4-yl)ethyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide, SULFATE ION, ... | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2012-05-25 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Flexibility of the Flap in the Active Site of BACE1 as Revealed by Crystal Structures and MD simulations
To be Published, 2012
|
|
3UQX
 
 | | Crystal structure of BACE1 with its inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1R)-1-(4-fluorophenyl)ethyl]-N'-[(2S,3S)-3-hydroxy-4-{4-[(1S)-1-hydroxyethyl]-1H-1,2,3-triazol-1-yl}-1-phenylbutan-2-yl]-5-[methyl(methylsulfonyl)amino]benzene-1,3-dicarboxamide, ... | | Authors: | Chen, T.T, Chen, W.Y, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Flexibility of the Flap in the Active Site of BACE1 as Revealed by Crystal Structures and MD simulations
to be published
|
|
6VI4
 
 | | Nanobody-Enabled Monitoring of Kappa Opioid Receptor States | | Descriptor: | (3R)-7-hydroxy-N-{(2S)-1-[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethylpiperidin-1-yl]-3-methylbutan-2-yl}-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, CHOLESTEROL, Kappa opioid receptor, ... | | Authors: | Che, T, Roth, B.L. | | Deposit date: | 2020-01-11 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanobody-enabled monitoring of kappa opioid receptor states.
Nat Commun, 11, 2020
|
|
8IBH
 
 | | Cep57 C-terminal domain | | Descriptor: | Centrosomal protein of 57 kDa | | Authors: | Chen, T, Yeh, H.-W, Cheng, H.-C. | | Deposit date: | 2023-02-10 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cep57 regulates human centrosomes through multivalent interactions.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7XGV
 
 | | Legionella glucosyltransferase | | Descriptor: | Lgt2 | | Authors: | Chen, T.T, Ouyang, S.Y. | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis for the Action Mechanism of Legionella Glycosyltransferase.
Small Struct, 2023
|
|
7XGX
 
 | | Glucosyltransferase | | Descriptor: | Lgt2, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Chen, T.T, Ouyang, S.Y. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural Basis for the Action Mechanism of Legionella Glycosyltransferase.
Small Struct, 2023
|
|
6TWN
 
 | | Crystal structure of Talin1 R7R8 in complex with CDK1 (206-223) | | Descriptor: | CHLORIDE ION, Cyclin-dependent kinase 1, GLYCEROL, ... | | Authors: | Zacharchenko, T, Muench, S.P, Goult, B.T. | | Deposit date: | 2020-01-13 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Talin mechanosensitivity is modulated by a direct interaction with cyclin-dependent kinase-1.
J.Biol.Chem., 297, 2021
|
|
6B73
 
 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | Descriptor: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | Authors: | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
3GK9
 
 | | Crystal structure of murine Ngb under Xe pressure | | Descriptor: | Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Moschetti, T, Mueller, U, Schultze, J, Brunori, M, Vallone, B. | | Deposit date: | 2009-03-10 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of neuroglobin at high Xe and Kr pressure reveals partial conservation of globin internal cavities.
Biophys. J., 97, 2009
|
|
5GGZ
 
 | | Crystal structure of novel inhibitor bound with Hsp90 | | Descriptor: | Heat shock protein HSP 90-alpha, [2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-(2-ethoxy-7,8-dihydro-5~{H}-pyrido[4,3-d]pyrimidin-6-yl)methanone | | Authors: | Chen, T.T, Li, J, Xu, Y.C. | | Deposit date: | 2016-06-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Novel Tetrahydropyrido[4,3-d]pyrimidines as Potent Inhibitors of Chaperone Heat Shock Protein 90
J. Med. Chem., 59, 2016
|
|
3GKT
 
 | | Crystal structure of murine neuroglobin under Kr pressure | | Descriptor: | KRYPTON, Neuroglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Moschetti, T, Mueller, U, Schultze, J, Brunori, M, Vallone, B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of neuroglobin at high Xe and Kr pressure reveals partial conservation of globin internal cavities.
Biophys. J., 97, 2009
|
|
4GHI
 
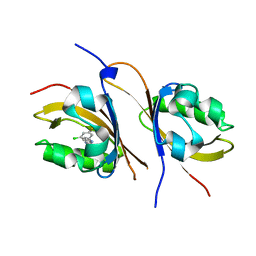 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains in complex with a benzoxadiazole antagonist | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, N-(3-chloro-5-fluorophenyl)-4-nitro-2,1,3-benzoxadiazol-5-amine | | Authors: | Scheuermann, T.H, Key, J, Tambar, U.K, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2012-08-07 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Allosteric inhibition of hypoxia inducible factor-2 with small molecules.
Nat.Chem.Biol., 9, 2013
|
|
3L9H
 
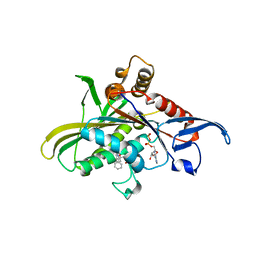 | | X-ray structure of mitotic kinesin-5 (KSP, KIF11, Eg5)in complex with the hexahydro-2H-pyrano[3,2-c]quinoline EMD 534085 | | Descriptor: | 1-[2-(dimethylamino)ethyl]-3-{[(2R,4aS,5R,10bS)-5-phenyl-9-(trifluoromethyl)-3,4,4a,5,6,10b-hexahydro-2H-pyrano[3,2-c]quinolin-2-yl]methyl}urea, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11 | | Authors: | Knoechel, T. | | Deposit date: | 2010-01-05 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The discovery and optimization of hexahydro-2H-pyrano[3,2-c]quinolines (HHPQs) as potent and selective inhibitors of the mitotic kinesin-5.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8OOI
 
 | | Full composite cryo-EM map of p97/VCP in ADP.Pi state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Cheng, T.C, Sakata, E, Schuetz, A.K. | | Deposit date: | 2023-04-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Characterizing ATP processing by the AAA+ protein p97 at the atomic level.
Nat.Chem., 16, 2024
|
|
8J9B
 
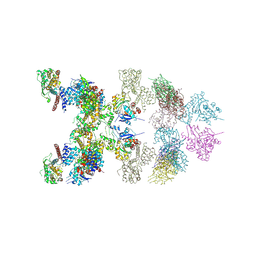 | | LnaB-actin binary complex | | Descriptor: | Actin gamma 1, Type IV secretion protein Dot | | Authors: | Chen, T.T, Ouyang, S.Y. | | Deposit date: | 2023-05-03 | | Release date: | 2024-05-08 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Legionella maintains host cell ubiquitin homeostasis by effectors with unique catalytic mechanisms.
Nat Commun, 15, 2024
|
|
8K6I
 
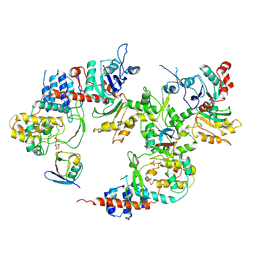 | | LnaB-Actin-PRUb ternary complex | | Descriptor: | Actin gamma 1, Legionella effector LnaB, PHOSPHATE ION, ... | | Authors: | Chen, T.T, Ouyang, S.Y. | | Deposit date: | 2023-07-25 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Complex structure of Legionella effector LnaB with host Actin and PR-Ub
To Be Published
|
|
8K6F
 
 | | LnaB-Actin-PRUb ternary complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin gamma 1, Legionella effector LnaB, ... | | Authors: | Chen, T.T, Ouyang, S.Y. | | Deposit date: | 2023-07-25 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Complex structure of Legionella effector LnaB with host Actin and PR-Ub
To Be Published
|
|
8K6R
 
 | |
8K6V
 
 | | LnaB-Actin-PRUb ternary complex | | Descriptor: | Actin gamma 1, LnaB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, T.T, Ouyang, S.Y. | | Deposit date: | 2023-07-25 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complex structure of Legionella effector LnaB with host Actin and ADPR-Ub
To Be Published
|
|
8K6E
 
 | | LnaB-Actin-PRUb ternary complex | | Descriptor: | Actin gamma 1, Legionella effector LnaB, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, T.T, Ouyang, S.Y. | | Deposit date: | 2023-07-25 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Complex structure of Legionella effector LnaB with host Actin and PR-Ub
To Be Published
|
|
8HCY
 
 | | Legionella glycosyltransferase | | Descriptor: | Dot/Icm secretion system substrate | | Authors: | Chen, T.T, Ouyang, S.Y. | | Deposit date: | 2022-11-03 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural Basis for the Action Mechanism of Legionella Glycosyltransferase.
Small Struct, 2023
|
|
6CZ1
 
 | | Crystal structure of ATPase domain of Human GRP78 bound to Ver155008 | | Descriptor: | 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, Endoplasmic reticulum chaperone BiP, MAGNESIUM ION | | Authors: | Antoshchenko, T, Chen, Y, Hughes, S, Park, H. | | Deposit date: | 2018-04-07 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystallographic selection of adenosine analogs that fit the mold of the active site of human GRP78 and beyond
To be Published
|
|
8Y1K
 
 | | The cryo-EM structure of TdpAB in complex with AMPPNP and PT-DNA | | Descriptor: | DNA (5'-D(P*GP*CP*CP*CP*TP*TP*TP*TP*GP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*CP*AP*AP*AP*AP*GP*GP*GP*C)-3'), PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, T.A, Qian, T. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A DNA phosphorothioation pathway via adenylated intermediate modulates Tdp machinery.
Nat.Chem.Biol., 2025
|
|
