2QZ6
 
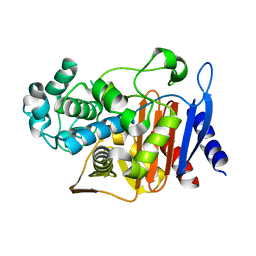 | | First crystal structure of a psychrophile class C beta-lactamase | | Descriptor: | Beta-lactamase | | Authors: | Michaux, C, Massant, J, Kerff, F, Charlier, P, Wouters, J. | | Deposit date: | 2007-08-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of a cold-adapted class C beta-lactamase
Febs J., 275, 2008
|
|
1ESU
 
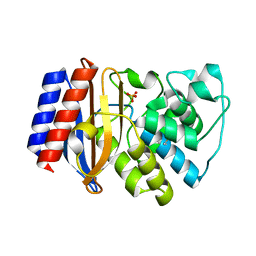 | | S235A MUTANT OF TEM1 BETA-LACTAMASE | | Descriptor: | BETA-LACTAMASE, SULFATE ION | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-11 | | Release date: | 2000-05-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TEM1 beta-lactamase structure solved by molecular replacement and refined structure of the S235A mutant.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
2WGV
 
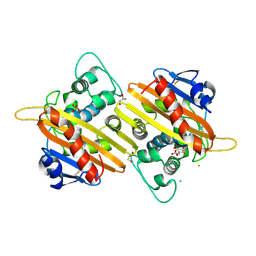 | | Crystal structure of the OXA-10 V117T mutant at pH 6.5 inhibited by a chloride ion | | Descriptor: | BETA-LACTAMASE OXA-10, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post- Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
1W7F
 
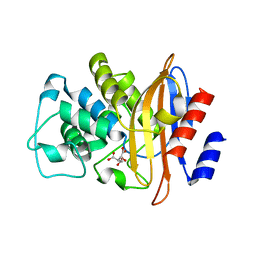 | |
2HPB
 
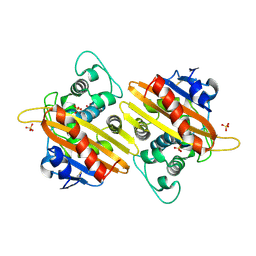 | | Crystal structure of the OXA-10 W154A mutant at pH 9.0 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HP6
 
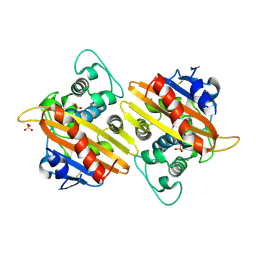 | | Crystal structure of the OXA-10 W154A mutant at pH 7.5 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2HP9
 
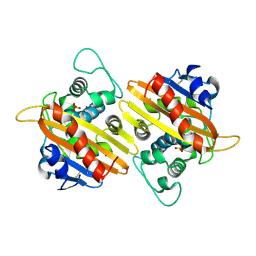 | | Crystal Structure of the OXA-10 W154A mutant at pH 6.0 | | Descriptor: | Beta-lactamase PSE-2, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2J9P
 
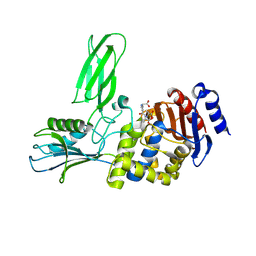 | | Crystal structure of the Bacillus subtilis PBP4a, and its complex with a peptidoglycan mimetic peptide. | | Descriptor: | (2R)-2-AMINO-7-{[(1R)-1-CARBOXYETHYL]AMINO}-7-OXOHEPTANOIC ACID, D-ALANINE, D-alanyl-D-alanine carboxypeptidase DacC | | Authors: | Sauvage, E, Herman, R, Kerff, F, Duez, C, Charlier, P. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Bacillus subtilis penicillin-binding protein 4a, and its complex with a peptidoglycan mimetic peptide.
J. Mol. Biol., 371, 2007
|
|
1HIX
 
 | | CRYSTALLOGRAPHIC ANALYSES OF FAMILY 11 ENDO-BETA-1,4-XYLANASE XYL1 FROM STREPTOMYCES SP. S38 | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Wouters, J, Georis, J, Dusart, J, Frere, J.M, Depiereux, E, Charlier, P. | | Deposit date: | 2001-01-05 | | Release date: | 2001-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Analysis of Family 11 Endo-[Beta]-1,4-Xylanase Xyl1 from Streptomyces Sp. S38
Acta Crystallogr.,Sect.D, 57, 2001
|
|
6G88
 
 | | Crystal structure of Enterococcus Faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyrrolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
6YCF
 
 | | Structure the bromelain protease from Ananas comosus in complex with the E64 inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, FBSB, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6YCG
 
 | | Structure the bromelain protease from Ananas comosus in complex with the TLCK inhibitor | | Descriptor: | CITRIC ACID, FBSB, ISOPROPYL ALCOHOL, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
1I2S
 
 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 | | Descriptor: | BETA-LACTAMASE, CITRIC ACID, SODIUM ION | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the
acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
6YCE
 
 | | Structure the bromelain protease from Ananas comosus with a thiomethylated active cysteine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FBSB, ISOPROPYL ALCOHOL, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
1I2W
 
 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 COMPLEXED WITH CEFOXITIN | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, BETA-LACTAMASE, CARBAMIC ACID | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
2BH0
 
 | | Crystal structure of a SeMet derivative of EXPA from Bacillus subtilis at 2.5 angstrom | | Descriptor: | YOAJ | | Authors: | Petrella, S, Herman, R, Sauvage, E, Filee, P, Joris, B, Charlier, P. | | Deposit date: | 2005-01-06 | | Release date: | 2006-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Activity of Bacillus Subtilis Yoaj (Exlx1), a Bacterial Expansin that Promotes Root Colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2RL3
 
 | | Crystal structure of the OXA-10 W154H mutant at pH 7 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase PSE-2, GLYCEROL, ... | | Authors: | Vercheval, L, Kerff, F, Herman, R, Sauvage, E, Guiet, R, Charlier, P, Frere, J.-M, Galleni, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2VGK
 
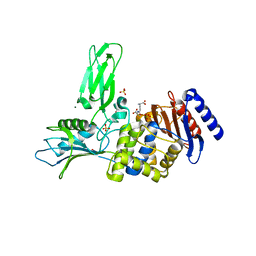 | | Crystal structure of Actinomadura R39 DD-peptidase complexed with a peptidoglycan-mimetic cephalosporin | | Descriptor: | (2R)-2-AMINO-7-{[(1R)-1-CARBOXYETHYL]AMINO}-7-OXOHEPTANOIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, kerff, F, Herman, R, Charlier, P. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of Complexes of Bacterial Dd-Peptidases with Peptidoglycan-Mimetic Ligands: The Substrate Specificity Puzzle.
J.Mol.Biol., 381, 2008
|
|
2VGJ
 
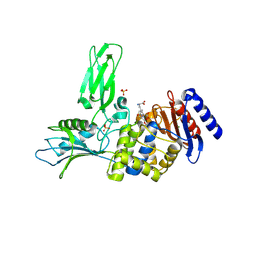 | | Crystal structure of Actinomadura R39 DD-peptidase complexed with a peptidoglycan-mimetic cephalosporin | | Descriptor: | CEPHALOSPORIN, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, kerff, F, Herman, R, Charlier, P. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Complexes of Bacterial Dd-Peptidases with Peptidoglycan-Mimetic Ligands: The Substrate Specificity Puzzle.
J.Mol.Biol., 381, 2008
|
|
6YCB
 
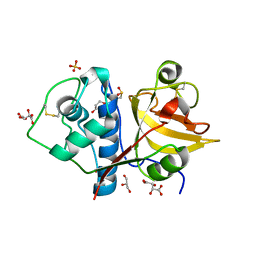 | | Structure the ananain protease from Ananas comosus covalently bound to with the E64 inhibitor | | Descriptor: | Ananain, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.257 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6G0K
 
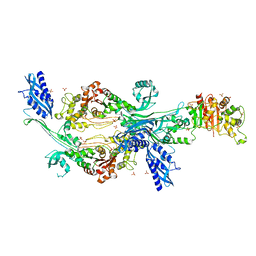 | | Crystal structure of Enterococcus faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-03-19 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
6YCD
 
 | | Structure the ananain protease from Ananas comosus covalently bound to the TLCK inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6YCC
 
 | | Structure the ananain protease from Ananas comosus covalently bound to the E64 inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
2HP5
 
 | | Crystal Structure of the OXA-10 W154G mutant at pH 7.0 | | Descriptor: | Beta-lactamase PSE-2, COBALT (II) ION, SULFATE ION | | Authors: | Kerff, F, Falzone, C, Herman, R, Sauvage, E, Charlier, P. | | Deposit date: | 2006-07-17 | | Release date: | 2007-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
1W5D
 
 | | Crystal structure of PBP4a from Bacillus subtilis | | Descriptor: | CALCIUM ION, PENICILLIN-BINDING PROTEIN | | Authors: | Sauvage, E, Herman, R, Petrella, S, Duez, C, Frere, J.M, Charlier, P. | | Deposit date: | 2004-08-06 | | Release date: | 2005-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Bacillus Subtilis Penicillin-Binding Protein 4A, and its Complex with a Peptidoglycan Mimetic Peptide.
J.Mol.Biol., 371, 2007
|
|
