1LBU
 
 | | HYDROLASE METALLO (ZN) DD-PEPTIDASE | | Descriptor: | MURAMOYL-PENTAPEPTIDE CARBOXYPEPTIDASE, ZINC ION | | Authors: | Charlier, P, Wery, J.-P, Dideberg, O, Frere, J.-M. | | Deposit date: | 1996-03-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Streptomyces Albus G D-Ala-A-Ala Carboxypeptidase
Handbook of Metalloproteins, 3, 2004
|
|
2J9P
 
 | | Crystal structure of the Bacillus subtilis PBP4a, and its complex with a peptidoglycan mimetic peptide. | | Descriptor: | (2R)-2-AMINO-7-{[(1R)-1-CARBOXYETHYL]AMINO}-7-OXOHEPTANOIC ACID, D-ALANINE, D-alanyl-D-alanine carboxypeptidase DacC | | Authors: | Sauvage, E, Herman, R, Kerff, F, Duez, C, Charlier, P. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Bacillus subtilis penicillin-binding protein 4a, and its complex with a peptidoglycan mimetic peptide.
J. Mol. Biol., 371, 2007
|
|
2VGK
 
 | | Crystal structure of Actinomadura R39 DD-peptidase complexed with a peptidoglycan-mimetic cephalosporin | | Descriptor: | (2R)-2-AMINO-7-{[(1R)-1-CARBOXYETHYL]AMINO}-7-OXOHEPTANOIC ACID, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, kerff, F, Herman, R, Charlier, P. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures of Complexes of Bacterial Dd-Peptidases with Peptidoglycan-Mimetic Ligands: The Substrate Specificity Puzzle.
J.Mol.Biol., 381, 2008
|
|
2VGJ
 
 | | Crystal structure of Actinomadura R39 DD-peptidase complexed with a peptidoglycan-mimetic cephalosporin | | Descriptor: | CEPHALOSPORIN, D-ALANYL-D-ALANINE CARBOXYPEPTIDASE, MAGNESIUM ION, ... | | Authors: | Sauvage, E, kerff, F, Herman, R, Charlier, P. | | Deposit date: | 2007-11-14 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Complexes of Bacterial Dd-Peptidases with Peptidoglycan-Mimetic Ligands: The Substrate Specificity Puzzle.
J.Mol.Biol., 381, 2008
|
|
7AP7
 
 | | Structure of the W64R amyloidogenic variant of human lysozyme | | Descriptor: | Lysozyme C, SULFATE ION | | Authors: | Vettore, N, Herman, R, Kerff, F, Charlier, P, Sauvage, E, Brans, A, Morray, J, Dobson, C, Kumita, J, Dumoulin, M. | | Deposit date: | 2020-10-16 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Characterisation of the structural, dynamic and aggregation properties of the W64R amyloidogenic variant of human lysozyme.
Biophys.Chem., 271, 2021
|
|
2WGV
 
 | | Crystal structure of the OXA-10 V117T mutant at pH 6.5 inhibited by a chloride ion | | Descriptor: | BETA-LACTAMASE OXA-10, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Vercheval, L, Kerff, F, Bauvois, C, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-04-27 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post- Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
3TVL
 
 | | Complex between the human thiamine triphosphatase and triphosphate | | Descriptor: | 1,2-ETHANEDIOL, TRIPHOSPHATE, Thiamine-triphosphatase | | Authors: | Delvaux, D, Herman, R, Sauvage, E, Wins, P, Bettendorff, L, Charlier, P, Kerff, F. | | Deposit date: | 2011-09-20 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of specificity and catalytic mechanism in mammalian 25-kDa thiamine triphosphatase.
Biochim.Biophys.Acta, 1830, 2013
|
|
4N1H
 
 | | Structure of a single-domain camelid antibody fragment cAb-F11N in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, Camelid heavy-chain antibody variable fragment cAb-F11N | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-10-04 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
5AEB
 
 | | Crystal structure of the class B3 di-zinc metallo-beta-lactamase LRA- 12 from an Alaskan soil metagenome. | | Descriptor: | COBALT (II) ION, LRA-12, SULFATE ION, ... | | Authors: | Power, P, Herman, R, Kerff, F, Bouillenne, F, Rodriguez, M.M, Galleni, M, Handelsman, J, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2015-08-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and kinetic analysis of the class B3 di-zinc metallo-beta-lactamase LRA-12 from an Alaskan soil metagenome.
PLoS ONE, 12, 2017
|
|
4M3K
 
 | | Structure of a single domain camelid antibody fragment cAb-H7S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, CHLORIDE ION, Camelid heavy-chain antibody variable fragment cAb-H7S | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
4M3J
 
 | | Structure of a single-domain camelid antibody fragment cAb-H7S specific of the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Camelid heavy-chain antibody variable fragment cAb-H7S, SULFATE ION | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
1J9M
 
 | | K38H mutant of Streptomyces K15 DD-transpeptidase | | Descriptor: | CHLORIDE ION, DD-transpeptidase, SODIUM ION | | Authors: | Fonze, E, Rhazi, N, Nguyen-Disteche, M, Charlier, P. | | Deposit date: | 2001-05-28 | | Release date: | 2001-06-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
1I2S
 
 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 | | Descriptor: | BETA-LACTAMASE, CITRIC ACID, SODIUM ION | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the
acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
1I2W
 
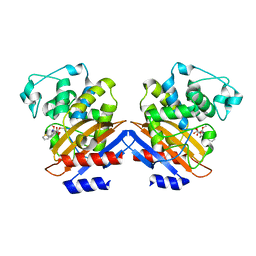 | | BETA-LACTAMASE FROM BACILLUS LICHENIFORMIS BS3 COMPLEXED WITH CEFOXITIN | | Descriptor: | (2R)-5-[(carbamoyloxy)methyl]-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, BETA-LACTAMASE, CARBAMIC ACID | | Authors: | Fonze, E, Vanhove, M, Dive, G, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-02-12 | | Release date: | 2002-03-13 | | Last modified: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the acyl-enzyme adduct formed with cefoxitin
Biochemistry, 41, 2002
|
|
6Y6L
 
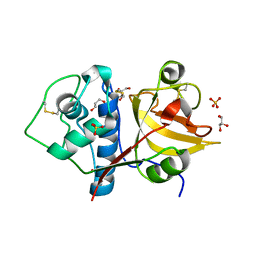 | | Structure the ananain protease from Ananas comosus with a thiomethylated catalytic cysteine | | Descriptor: | Ananain, GLYCEROL, SULFATE ION | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-02-26 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
2RL3
 
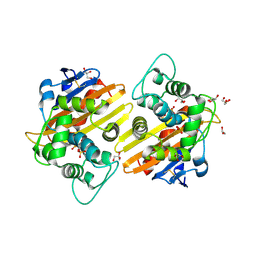 | | Crystal structure of the OXA-10 W154H mutant at pH 7 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase PSE-2, GLYCEROL, ... | | Authors: | Vercheval, L, Kerff, F, Herman, R, Sauvage, E, Guiet, R, Charlier, P, Frere, J.-M, Galleni, M. | | Deposit date: | 2007-10-18 | | Release date: | 2008-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Critical role of tryptophan 154 for the activity and stability of class D beta-lactamases.
Biochemistry, 48, 2009
|
|
2QZ6
 
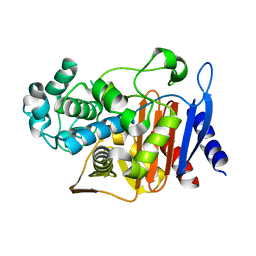 | | First crystal structure of a psychrophile class C beta-lactamase | | Descriptor: | Beta-lactamase | | Authors: | Michaux, C, Massant, J, Kerff, F, Charlier, P, Wouters, J. | | Deposit date: | 2007-08-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of a cold-adapted class C beta-lactamase
Febs J., 275, 2008
|
|
6YCB
 
 | | Structure the ananain protease from Ananas comosus covalently bound to with the E64 inhibitor | | Descriptor: | Ananain, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.257 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
1W7F
 
 | |
6YCE
 
 | | Structure the bromelain protease from Ananas comosus with a thiomethylated active cysteine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FBSB, ISOPROPYL ALCOHOL, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6YCF
 
 | | Structure the bromelain protease from Ananas comosus in complex with the E64 inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, FBSB, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6YCD
 
 | | Structure the ananain protease from Ananas comosus covalently bound to the TLCK inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6YCC
 
 | | Structure the ananain protease from Ananas comosus covalently bound to the E64 inhibitor | | Descriptor: | Ananain, GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
2BH0
 
 | | Crystal structure of a SeMet derivative of EXPA from Bacillus subtilis at 2.5 angstrom | | Descriptor: | YOAJ | | Authors: | Petrella, S, Herman, R, Sauvage, E, Filee, P, Joris, B, Charlier, P. | | Deposit date: | 2005-01-06 | | Release date: | 2006-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Activity of Bacillus Subtilis Yoaj (Exlx1), a Bacterial Expansin that Promotes Root Colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1SKF
 
 | | CRYSTAL STRUCTURE OF THE STREPTOMYCES K15 DD-TRANSPEPTIDASE | | Descriptor: | D-ALANYL-D-ALANINE TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 1998-08-20 | | Release date: | 1999-08-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a penicilloyl-serine transferase of intermediate penicillin sensitivity. The DD-transpeptidase of streptomyces K15.
J.Biol.Chem., 274, 1999
|
|
