1XG7
 
 | | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus | | Descriptor: | hypothetical protein | | Authors: | Chang, J, Zhao, M, Horanyi, P, Xu, H, Yang, H, Liu, Z.-J, Chen, L, Zhou, W, Habel, J, Tempel, W, Lee, D, Lin, D, Chang, S.-H, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.-S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Chen, C.-Y, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Conserved hypothetical protein Pfu-877259-001 from Pyrococcus furiosus
To be published
|
|
1ZGH
 
 | | Methionyl-tRNA formyltransferase from Clostridium thermocellum | | Descriptor: | Methionyl-tRNA formyltransferase, UNKNOWN ATOM OR ION | | Authors: | Yang, H, Kataeva, I, Xu, H, Zhao, M, Chang, J, Liu, Z, Chen, L, Tempel, W, Habel, J, Zhou, W, Lee, D, Lin, D, Chang, S, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Wang, B, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-21 | | Release date: | 2005-05-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Methionyl-tRNA formyltransferase from Clostridium thermocellum
To be published
|
|
3K6G
 
 | | Crystal structure of Rap1 and TRF2 complex | | Descriptor: | Telomeric repeat-binding factor 2, Telomeric repeat-binding factor 2-interacting protein 1 | | Authors: | Chen, Y, Rai, R, Yang, Y.T, Zheng, H, Chang, S, Lei, M. | | Deposit date: | 2009-10-08 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A conserved motif within RAP1 has diversified roles in telomere protection and regulation in different organisms.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1ZTD
 
 | | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus | | Descriptor: | Hypothetical Protein Pfu-631545-001 | | Authors: | Fu, Z.-Q, Horanyi, P, Florence, Q, Liu, Z.-J, Chen, L, Lee, D, Habel, J, Xu, H, Nguyen, D, Chang, S.-H, Zhou, W, Zhang, H, Jenney Jr, F.E, Sha, B, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus
To be Published
|
|
7XUM
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with Cu+ in presence of ATOX1 | | Descriptor: | COPPER (II) ION, Copper-transporting ATPase 2 | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
7XUO
 
 | | Structure of ATP7B C983S/C985S/D1027A mutant with cisplatin in presence of ATOX1 | | Descriptor: | Copper-transporting ATPase 2, PLATINUM (II) ION | | Authors: | Yang, G, Xu, L, Chang, S, Guo, J, Wu, Z. | | Deposit date: | 2022-05-19 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the human Wilson disease copper transporter ATP7B.
Cell Rep, 42, 2023
|
|
3E5R
 
 | |
3E6A
 
 | |
8I51
 
 | | Acyl-ACP synthetase structure bound to AMP-MC7 | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-21 | | Release date: | 2024-01-24 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-MC7
To Be Published
|
|
8I6M
 
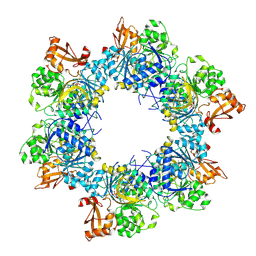 | | Acyl-ACP synthetase structure bound to AMP-C18:1 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-acyl carrier protein synthetase, MAGNESIUM ION, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-01-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Acyl-ACP synthetase structure bound to AMP-C18:1
To Be Published
|
|
4QFI
 
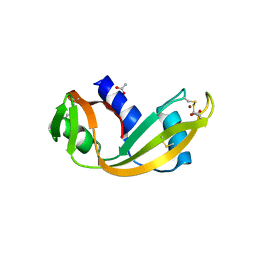 | | The crystal structure of rat angiogenin-heparin complex | | Descriptor: | ACETIC ACID, Angiogenin, ZINC ION | | Authors: | Yeo, K.J, Hwang, E, Min, K.M, Hwang, K.Y, Jeon, Y.H, Chang, S.I, Cheong, H.K. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | The crystal structure of rat angiogenin-heparin complex
To be Published
|
|
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | Descriptor: | Pachytene checkpoint protein 2 homolog | | Authors: | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | Deposit date: | 2019-12-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
3V1Y
 
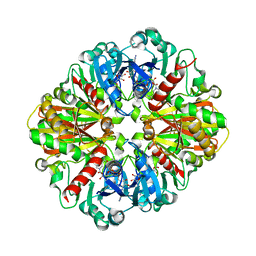 | | Crystal structures of glyceraldehyde-3-phosphate dehydrogenase complexes with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tien, Y.C, Chuankhayan, P, Lin, Y.H, Chang, S.L, Chen, C.J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis
Plant Mol.Biol., 80, 2012
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
8I8D
 
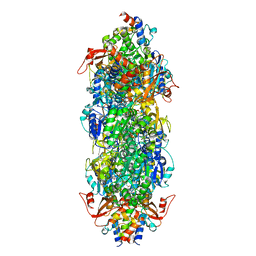 | | Acyl-ACP synthetase structure bound to MC7-ACP | | Descriptor: | 7-methoxy-7-oxidanylidene-heptanoic acid, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Acyl-ACP synthetase structure bound to MC7-ACP
To Be Published
|
|
8I8E
 
 | | Acyl-ACP synthetase structure bound to C18:1-ACP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, ADENOSINE MONOPHOSPHATE, Acyl carrier protein, ... | | Authors: | Huang, H, Wang, C, Chang, S, Cui, T, Xu, Y, Zhang, H, Zhou, C, Zhang, X, Feng, Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Acyl-ACP synthetase structure bound to C18:1-ACP
To Be Published
|
|
1XHO
 
 | | Chorismate mutase from Clostridium thermocellum Cth-682 | | Descriptor: | Chorismate mutase, UNKNOWN ATOM OR ION | | Authors: | Xu, H, Chen, L, Lee, D, Habel, J.E, Nguyen, J, Chang, S.-H, Kataeva, I, Chang, J, Zhao, M, Yang, H, Horanyi, P, Florence, Q, Tempel, W, Zhou, W, Lin, D, Zhang, H, Praissman, J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Ljungdahl, L, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-20 | | Release date: | 2004-11-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Away from the edge II: in-house Se-SAS phasing with chromium radiation.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZD0
 
 | | Crystal structure of Pfu-542154 conserved hypothetical protein | | Descriptor: | MAGNESIUM ION, METHANOL, UNKNOWN ATOM OR ION, ... | | Authors: | Habel, J.E, Liu, Z.J, Horanyi, P.S, Florence, Q.J.T, Tempel, W, Zhou, W, Chen, L, Lee, D, Nguyen, J, Chang, S.H, Bereton, P, Izumi, M, Jenny Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-04-13 | | Release date: | 2005-05-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Pfu-542154 conserved hypothetical protein
To be Published
|
|
1Y82
 
 | | Conserved hypothetical protein Pfu-367848-001 from Pyrococcus furiosus | | Descriptor: | UNKNOWN ATOM OR ION, hypothetical protein | | Authors: | Horanyi, P, Tempel, W, Habel, J, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Florence, Q, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-367848-001 from Pyrococcus furiosus
To be published
|
|
4QFJ
 
 | | The crystal structure of rat angiogenin-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACETIC ACID, Angiogenin, ... | | Authors: | Yeo, K.J, Hwang, E, Min, K.M, Hwang, K.Y, Jeon, Y.H, Chang, S.I, Cheong, H.K. | | Deposit date: | 2014-05-21 | | Release date: | 2014-08-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | The crystal structure of rat angiogenin-heparin complex
To be Published
|
|
5IV8
 
 | | The LPS Transporter LptDE from Klebsiella pneumoniae, core complex | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LPS biosynthesis protein, LPS-assembly lipoprotein LptE | | Authors: | Botos, I, McCarthy, J.G, Buchanan, S.K. | | Deposit date: | 2016-03-20 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.938 Å) | | Cite: | Structural and Functional Characterization of the LPS Transporter LptDE from Gram-Negative Pathogens.
Structure, 24, 2016
|
|
3C3M
 
 | | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Dickey, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1.
To be Published
|
|
