3B6Y
 
 | | Crystal Structure of the Second HIN-200 Domain of Interferon-Inducible Protein 16 | | Descriptor: | Gamma-interferon-inducible protein Ifi-16, SULFATE ION | | Authors: | Liao, J.C.C, Lam, R, Ravichandran, M, Duan, S, Tempel, W, Chirgadze, N.Y, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-29 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure Analysis of the Second HIN Domain of IFI16.
To be Published
|
|
3C5K
 
 | | Crystal structure of human HDAC6 zinc finger domain | | Descriptor: | Histone deacetylase 6, ZINC ION | | Authors: | Dong, A, Ravichandran, M, Schuetz, A, Loppnau, P, Li, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Dhe-Paganon, S, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human HDAC6 zinc finger domain.
To be Published
|
|
4EDU
 
 | | The MBT repeats of human SCML2 in a complex with histone H2A peptide | | Descriptor: | Histone H2A.J peptide, Sex comb on midleg-like protein 2 | | Authors: | Nady, N, Amaya, M.F, Tempel, W, Ravichandran, M, Arrowsmith, C.H. | | Deposit date: | 2012-03-27 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Histone recognition by human malignant brain tumor domains.
J.Mol.Biol., 423, 2012
|
|
3DM1
 
 | | Crystal structure of the complex of human chromobox homolog 3 (CBX3) with peptide | | Descriptor: | Chromobox protein homolog 3, Histone-lysine N-methyltransferase, H3 lysine-9 specific 3 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
4ER8
 
 | | Structure of the REP associates tyrosine transposase bound to a REP hairpin | | Descriptor: | DNA (32-MER), NICKEL (II) ION, TnpArep for protein | | Authors: | Messing, S.A.J, Ton-Hoang, B, Hickman, A.B, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2012-04-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The processing of repetitive extragenic palindromes: the structure of a repetitive extragenic palindrome bound to its associated nuclease.
Nucleic Acids Res., 40, 2012
|
|
3GV4
 
 | | Crystal structure of human HDAC6 zinc finger domain and ubiquitin C-terminal peptide RLRGG | | Descriptor: | CALCIUM ION, Histone deacetylase 6, ZINC ION, ... | | Authors: | Dong, A, Ravichandran, M, Loppnau, P, Li, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Dhe-Paganon, S, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of human HDAC6 zinc finger domain and ubiquitin C-terminal peptide RLRGG
To be Published
|
|
2A6O
 
 | | Crystal Structure of the ISHp608 Transposase in Complex with Stem-loop DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*TP*AP*GP*CP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*GP*A)-3', ISHp608 Transposase | | Authors: | Ronning, D.R, Guynet, C, Ton-Hoang, B, Perez, Z.N, Ghirlando, R, Chandler, M, Dyda, F. | | Deposit date: | 2005-07-03 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Active site sharing and subterminal hairpin recognition in a new class of DNA transposases.
Mol.Cell, 20, 2005
|
|
3TZD
 
 | | Crystal structure of the complex of Human Chromobox Homolog 3 (CBX3) | | Descriptor: | Chromobox protein homolog 3, Histone H1.4 | | Authors: | Amaya, M.F, Ravichandran, M, Loppnau, P, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-09-27 | | Release date: | 2012-03-07 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a.
Plos One, 7, 2012
|
|
1AP4
 
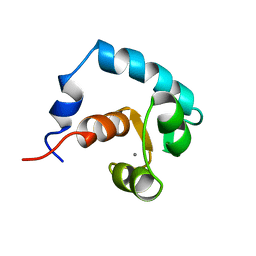 | | REGULATORY DOMAIN OF HUMAN CARDIAC TROPONIN C IN THE CALCIUM-SATURATED STATE, NMR, 40 STRUCTURES | | Descriptor: | CALCIUM ION, CARDIAC N-TROPONIN C | | Authors: | Li, M.X, Spyracopoulos, L, Sia, S.K, Gagne, S.M, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 1997-07-24 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural transition in the regulatory domain of human cardiac troponin C.
Biochemistry, 36, 1997
|
|
1EJJ
 
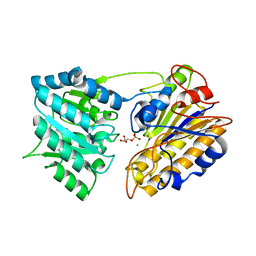 | | CRYSTAL STRUCTURAL ANALYSIS OF PHOSPHOGLYCERATE MUTASE COCRYSTALLIZED WITH 3-PHOSPHOGLYCERATE | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, PHOSPHOGLYCERATE MUTASE | | Authors: | Jedrzejas, M.J, Chander, M, Setlow, P, Krishnasamy, G. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of action of a novel phosphoglycerate mutase from Bacillus stearothermophilus.
EMBO J., 19, 2000
|
|
3FM0
 
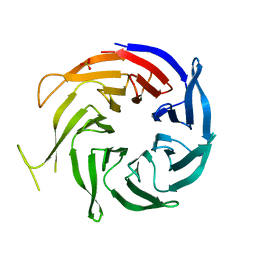 | | Crystal structure of WD40 protein Ciao1 | | Descriptor: | Protein CIAO1, SULFATE ION | | Authors: | Dong, A, Ravichandran, M, Crombet, L, Cossar, D, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Bountra, C, Bochkarev, A, Min, J, Ouyang, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
2K1B
 
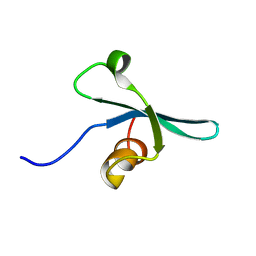 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 7 | | Descriptor: | Chromobox protein homolog 7 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Gutmanas, A, Fares, C, Bountra, C, Weigelt, J, Loppnau, P, Ravichandran, M, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-25 | | Release date: | 2008-03-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromobox protein homolog 7.
To be Published
|
|
3K35
 
 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
4FL6
 
 | | Crystal structure of the complex of the 3-MBT repeat domain of L3MBTL3 and UNC1215 | | Descriptor: | Lethal(3)malignant brain tumor-like protein 3, UNKNOWN ATOM OR ION, [2-(phenylamino)benzene-1,4-diyl]bis{[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone} | | Authors: | Zhong, N, Tempel, W, Ravichandran, M, Dong, A, Ingerman, L.A, Graslund, S, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain.
Nat. Chem. Biol., 9, 2013
|
|
2K28
 
 | | Solution NMR structure of the chromo domain of the chromobox protein homolog 4 | | Descriptor: | E3 SUMO-protein ligase CBX4 | | Authors: | Kaustov, L, Lemak, A, Quyang, H, Fares, C, Gutmanas, A, Ravichandran, M, Loppnau, P, Bountra, C, Weigelt, J, Edwards, A.M, Min, J, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the chromo domain of the chromobox protein homolog 4.
To be Published
|
|
1EQJ
 
 | | CRYSTAL STRUCTURE OF PHOSPHOGLYCERATE MUTASE FROM BACILLUS STEAROTHERMOPHILUS COMPLEXED WITH 2-PHOSPHOGLYCERATE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, MANGANESE (II) ION, PHOSPHOGLYCERATE MUTASE | | Authors: | Jedrzejas, M.J, Chander, M, Setlow, P, Krishnasamy, G. | | Deposit date: | 2000-04-05 | | Release date: | 2001-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of catalysis of the cofactor-independent phosphoglycerate mutase from Bacillus stearothermophilus. Crystal structure of the complex with 2-phosphoglycerate.
J.Biol.Chem., 275, 2000
|
|
2VJV
 
 | | Crystal structure of the IS608 transposase in complex with left end 26-mer DNA hairpin and a 6-mer DNA representing the left end cleavage site | | Descriptor: | 5'-D(*DA*DA*DA*DG*DC*DC*DC*DC*DT*DA*DG*DC*DTP*DT *DT*DT*DA*DG*DC*DT*DA*DT*DG*DG*DG*DGP)-3', 5'-D(*DT*DA*DT*DT*DA*DCP)-3', MAGNESIUM ION, ... | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2VIC
 
 | | CRYSTAL STRUCTURE OF THE ISHP608 TRANSPOSASE IN COMPLEX with Left end 26- mer DNA and manganese | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', MANGANESE (II) ION, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2XM3
 
 | | Deinococcus radiodurans ISDra2 Transposase Left end DNA complex | | Descriptor: | 5'-D(*TP*TP*AP*GP*T)-3', ACETATE ION, DRA2 TRANSPOSASE BINDING ELEMENT, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-22 | | Release date: | 2010-10-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2OWA
 
 | | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040) | | Descriptor: | Arfgap-like finger domain containing protein, ZINC ION | | Authors: | Dong, A, Lew, J, Zhao, Y, Hassanali, A, Lin, L, Ravichandran, M, Wasney, G, Vedadi, M, Kozieradzki, I, Bochkarev, A, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Hui, R, Qiu, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-02-15 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative GTPase activating protein for ADP ribosylation factor from Cryptosporidium parvum (cgd5_1040)
To be Published
|
|
2VHG
 
 | | Crystal Structure of the ISHp608 Transposase in Complex with Right End 31-mer DNA | | Descriptor: | MANGANESE (II) ION, RIGHT END 31-MER, TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-11-21 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
2XO6
 
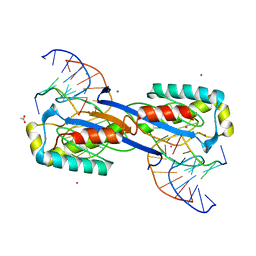 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE Y132F MUTANT COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE | | Descriptor: | 5'-D(*TP*TP*GP*AP*TP*G)-3', ACETATE ION, CADMIUM ION, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-08-09 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2XQC
 
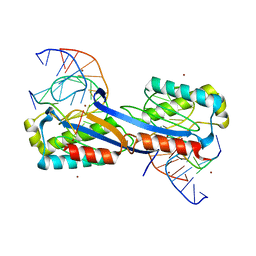 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE AND ZN | | Descriptor: | 5'-D(TP*TP*GP*AP*TP*GP)-3', DRA2 TRANSPOSASE LEFT END RECOGNITION SEQUENCE, TRANSPOSASE, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-09-01 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2XMA
 
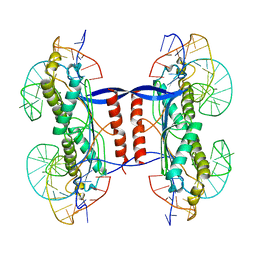 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE RIGHT END DNA COMPLEX | | Descriptor: | DRA2 TRANSPOSASE RIGHT END RECOGNITION SITE, MAGNESIUM ION, TRANSPOSASE | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-07-26 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
2VIH
 
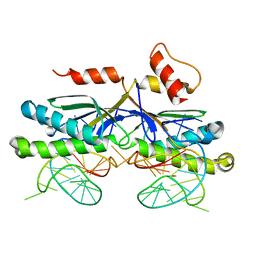 | | CRYSTAL STRUCTURE OF THE IS608 TRANSPOSASE IN COMPLEX WITH Left END 26-MER DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*CP*CP*CP*CP*TP*AP *GP*CP*TP*TP*TP*TP*AP*GP*CP*TP*AP*TP*GP*GP*GP*G)-3', TRANSPOSASE ORFA | | Authors: | Barabas, O, Ronning, D.R, Guynet, C, Hickman, A.B, Ton-Hoang, B, Chandler, M, Dyda, F. | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of is200/is605 Family DNA Transposases: Activation and Transposon-Directed Target Site Selection.
Cell(Cambridge,Mass.), 132, 2008
|
|
