5UG1
 
 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain with methylsulfonyl adduct | | Descriptor: | Acyltransferase, SODIUM ION, methanesulfonic acid | | Authors: | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-10-25 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
5UFY
 
 | | Structure of Streptococcus pneumoniae peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acyltransferase, SODIUM ION | | Authors: | Sychantha, D, Jones, C, Little, D.J, Moynihan, P.J, Robinson, H, Galley, N.F, Roper, D.I, Dowson, C.G, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-01-06 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | In vitro characterization of the antivirulence target of Gram-positive pathogens, peptidoglycan O-acetyltransferase A (OatA).
PLoS Pathog., 13, 2017
|
|
5V8E
 
 | | Structure of Bacillus cereus PatB1 | | Descriptor: | Bacillus cereus PatB1, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sychantha, D, Little, D.J, Chapman, R.N, Boons, G.J, Robinson, H, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-03-21 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PatB1 is an O-acetyltransferase that decorates secondary cell wall polysaccharides.
Nat. Chem. Biol., 14, 2018
|
|
8SPM
 
 | | Crystal structure of NikA in complex Ni-AMA | | Descriptor: | Aspergillomarasmine A, NICKEL (II) ION, Nickel ABC transporter, ... | | Authors: | Sychantha, D, Prehna, G, Wright, G.D. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Targeting bacterial nickel transport with aspergillomarasmine A suppresses virulence-associated Ni-dependent enzymes.
Nat Commun, 15, 2024
|
|
5V8D
 
 | | Structure of Bacillus cereus PatB1 with sulfonyl adduct | | Descriptor: | Bacillus cereus PatB1, SULFATE ION | | Authors: | Sychantha, D, Little, D.J, Chapman, R.N, Boons, G.J, Robinson, H, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-03-21 | | Release date: | 2017-10-18 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | PatB1 is an O-acetyltransferase that decorates secondary cell wall polysaccharides.
Nat. Chem. Biol., 14, 2018
|
|
8UK6
 
 | |
6BT4
 
 | | Crystal structure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit | | Descriptor: | 2-(acetylamino)-4-O-{2-(acetylamino)-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranose, S-layer protein sap, SULFATE ION | | Authors: | Sychantha, D, Chapman, R.N, Bamford, N.C, Boons, G.J, Howell, P.L, Clarke, A.J. | | Deposit date: | 2017-12-05 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Molecular Basis for the Attachment of S-Layer Proteins to the Cell Wall of Bacillus anthracis.
Biochemistry, 57, 2018
|
|
5MWO
 
 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7E8. | | Descriptor: | (5-methyl-1-benzothiophen-2-yl)methanol, 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MXK
 
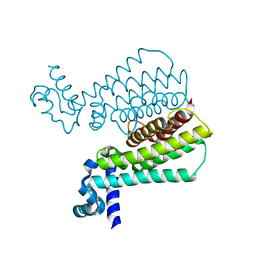 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7G9. | | Descriptor: | 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR, ~{N}-(5-oxidanylidene-7,8-dihydro-6~{H}-naphthalen-2-yl)ethanamide | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
1NLI
 
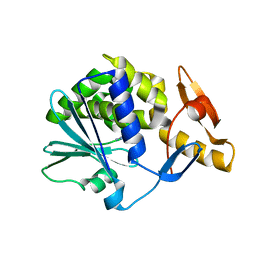 | | Complex of [E160A-E189A] trichosanthin and adenine | | Descriptor: | ADENINE, Ribosome-inactivating protein alpha-trichosanthin | | Authors: | Shaw, P.C, Wong, K.B, Chan, D.S.B, Williams, R.L. | | Deposit date: | 2003-01-07 | | Release date: | 2003-01-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for the interaction of [E160A-E189A]-trichosanthin with adenine.
Toxicon, 41, 2003
|
|
5EDT
 
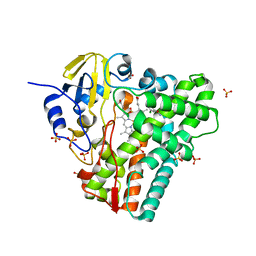 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with LIG9 | | Descriptor: | 2-azanyl-5-chloranyl-benzamide, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kavanagh, M.E, Coyne, A.G, McLean, K.J, James, G.G, Levy, C, Marino, L.B, Carvalho, L.P.D, Chan, D.S.H, Hudson, S.A, Surade, S, Munro, A.W, Abell, C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Fragment-Based Approaches to the Development of Mycobacterium tuberculosis CYP121 Inhibitors.
J.Med.Chem., 59, 2016
|
|
2W1O
 
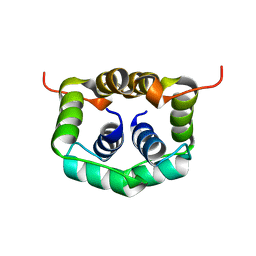 | | NMR structure of dimerization domain of human ribosomal protein P2 | | Descriptor: | 60S ACIDIC RIBOSOMAL PROTEIN P2 | | Authors: | Lee, K.M, Chan, D.S, Sze, K.H, Zhu, G, Shaw, P.C, Wong, K.B. | | Deposit date: | 2008-10-20 | | Release date: | 2009-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Dimerization Domain of Ribosomal Protein P2 Provides Insights for the Structural Organization of Eukaryotic Stalk.
Nucleic Acids Res., 38, 2010
|
|
4WL3
 
 | |
8G8W
 
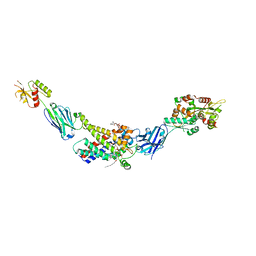 | | Molecular mechanism of nucleotide inhibition of human uncoupling protein 1 | | Descriptor: | CARDIOLIPIN, GUANOSINE-5'-TRIPHOSPHATE, Mitochondrial brown fat uncoupling protein 1, ... | | Authors: | Gogoi, P, Jones, S.A, Ruprecht, J.J, King, M.S, Lee, Y, Zogg, T, Pardon, E, Chand, D, Steimle, S, Copeman, D, Cotrim, C.A, Steyaert, J, Crichton, P.G, Moiseenkova-Bell, V, Kunji, E.R.S. | | Deposit date: | 2023-02-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of purine nucleotide inhibition of human uncoupling protein 1.
Sci Adv, 9, 2023
|
|
6VJP
 
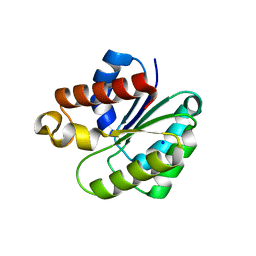 | | Structure of Staphylococcus aureus peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acetyltransferase, SODIUM ION | | Authors: | Jones, C.J, Sychantha, D, Howell, P.L, Clarke, A.J. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Structural basis for theO-acetyltransferase function of the extracytoplasmic domain of OatA fromStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
6WN9
 
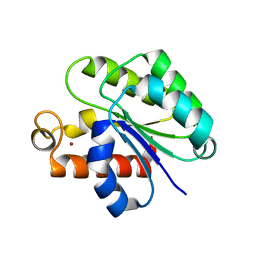 | | Structure of Staphylococcus aureus peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain, Zn-bound | | Descriptor: | Acetyltransferase, ZINC ION | | Authors: | Jones, C.J, Sychantha, D, Howell, P.L, Clarke, A.J. | | Deposit date: | 2020-04-22 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for theO-acetyltransferase function of the extracytoplasmic domain of OatA fromStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
7MK5
 
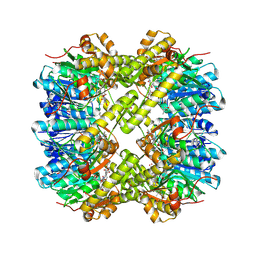 | | Crystal structure of Escherichia coli ClpP covalently inhibited by clipibicyclene | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-[(1E)-3-{[(2E,4E,6E,8S)-8-hydroxy-4-methyldeca-2,4,6-trienoyl]amino}-3-oxoprop-1-en-1-yl]azete-1(2H)-carboxylic acid, ACETATE ION, ... | | Authors: | Culp, E.J, Sychantha, D, Hobson, C, Pawlowski, A.J, Prehna, G, Wright, G.D. | | Deposit date: | 2021-04-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | ClpP inhibitors are produced by a widespread family of bacterial gene clusters.
Nat Microbiol, 7, 2022
|
|
5K5S
 
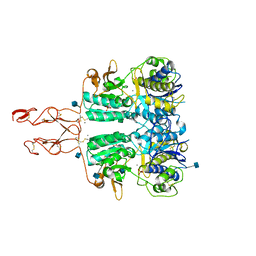 | | Crystal structure of the active form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
5K5T
 
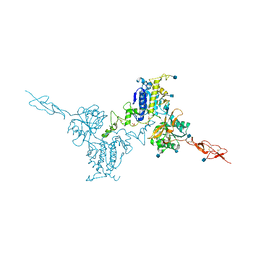 | | Crystal structure of the inactive form of human calcium-sensing receptor extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular calcium-sensing receptor, ... | | Authors: | Geng, Y, Mosyak, L, Kurinov, I, Zuo, H, Sturchler, E, Cheng, T.C, Subramanyam, P, Brown, A.P, Brennan, S.C, Mun, H.-C, Bush, M, Chen, Y, Nguyen, T, Cao, B, Chang, D, Quick, M, Conigrave, A, Colecraft, H.M, McDonald, P, Fan, Q.R. | | Deposit date: | 2016-05-23 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanism of ligand activation in human calcium-sensing receptor.
Elife, 5, 2016
|
|
4PEL
 
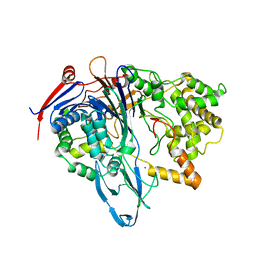 | | S1C mutant of Penicillin G acylase from Kluyvera citrophila | | Descriptor: | CALCIUM ION, Penicillin G acylase subunit alpha, Penicillin G acylase subunit beta | | Authors: | Ramasamy, S, Chand, D, Varshney, N.K, Brannigan, J.A, Wilkinson, A.J, Suresh, C.G. | | Deposit date: | 2014-04-24 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Penicillin G acylase
To Be Published
|
|
