6EO1
 
 | | The electron crystallography structure of the cAMP-bound potassium channel MloK1 (PCO-refined) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Kowal, J, Biyani, N, Chami, M, Scherer, S, Rzepiela, A, Baumgartner, P, Upadhyay, V, Nimigean, C, Stahlberg, H. | | Deposit date: | 2017-10-08 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON CRYSTALLOGRAPHY (4.5 Å) | | Cite: | High-Resolution Cryoelectron Microscopy Structure of the Cyclic Nucleotide-Modulated Potassium Channel MloK1 in a Lipid Bilayer.
Structure, 26, 2018
|
|
6YE4
 
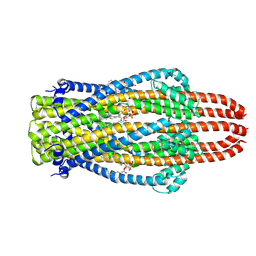 | | Structure of ExbB pentamer from Serratia marcescens by single particle cryo electron microscopy | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Biopolymer transport protein ExbB | | Authors: | Biou, V, Delepelaire, P, Coureux, P.D, Chami, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and molecular determinants for the interaction of ExbB from Serratia marcescens and HasB, a TonB paralog.
Commun Biol, 5, 2022
|
|
6TY3
 
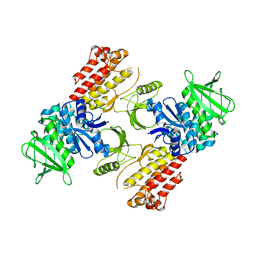 | | FAK structure from single particle analysis of 2D crystals | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Acebron, I, Righetto, R, Biyani, N, Chami, M, Boskovic, J, Stahlberg, H, Lietha, D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.32 Å) | | Cite: | Structural basis of Focal Adhesion Kinase activation on lipid membranes.
Embo J., 39, 2020
|
|
6TY4
 
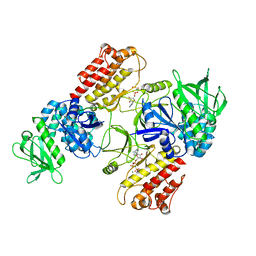 | | FAK structure with AMP-PNP from single particle analysis of 2D crystals | | Descriptor: | Focal adhesion kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Acebron, I, Righetto, R, Biyani, N, Chami, M, Boskovic, J, Stahlberg, H, Lietha, D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.96 Å) | | Cite: | Structural basis of Focal Adhesion Kinase activation on lipid membranes.
Embo J., 39, 2020
|
|
7ZNQ
 
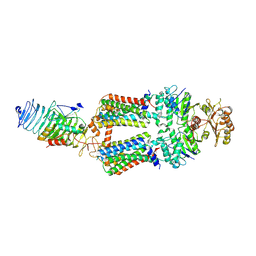 | | ABC transporter complex NosDFYL in GDN | | Descriptor: | COPPER (II) ION, Copper-binding lipoprotein NosL, MAGNESIUM ION, ... | | Authors: | Zhang, L, Mueller, C, Zipfel, S, Chami, M, Einsle, O. | | Deposit date: | 2022-04-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7AJQ
 
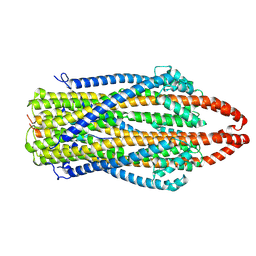 | | cryo-EM structure of ExbBD from Serratia Marcescens | | Descriptor: | Biopolymer transport protein ExbB, Biopolymer transport protein ExbD | | Authors: | Biou, V, Adaixo, R, Coureux, P.D, Delepelaire, P, Chami, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and molecular determinants for the interaction of ExbB from Serratia marcescens and HasB, a TonB paralog.
Commun Biol, 5, 2022
|
|
7AA5
 
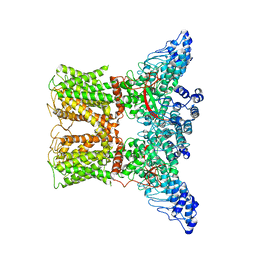 | | Human TRPV4 structure in presence of 4a-PDD | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 4,Green fluorescent protein | | Authors: | Botte, M, Ulrich, A.K.G, Adaixo, R, Gnutt, D, Brockmann, A, Bucher, D, Chami, M, Bocquet, M, Ebbinghaus-Kintscher, U, Puetter, V, Becker, A, Egner, U, Stahlberg, H, Hennig, M, Holton, S.J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structural studies of the agonist complexed human TRPV4 ion-channel reveals novel structural rearrangements resulting in an open-conformation
To Be Published
|
|
2N1F
 
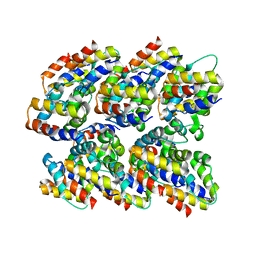 | | Structure and assembly of the mouse ASC filament by combined NMR spectroscopy and cryo-electron microscopy | | Descriptor: | Apoptosis-associated speck-like protein | | Authors: | Sborgi, L, Ravotti, F, Dandey, V, Dick, M, Mazur, A, Reckel, S, Chami, M, Scherer, S, Bockmann, A, Egelman, E, Stahlberg, H, Broz, P, Meier, B, Hiller, S. | | Deposit date: | 2015-04-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4CHW
 
 | | The electron crystallography structure of the cAMP-free potassium channel MloK1 | | Descriptor: | CYCLIC NUCLEOTIDE-GATED POTASSIUM CHANNEL MLL3241, POTASSIUM ION | | Authors: | Kowal, J, Chami, M, Baumgartner, P, Arheit, M, Chiu, P.L, Rangl, M, Scheuring, S, Schroeder, G.F, Nimigean, C.M, Stahlberg, H. | | Deposit date: | 2013-12-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Ligand-induced structural changes in the cyclic nucleotide-modulated potassium channel MloK1.
Nat Commun, 5, 2014
|
|
4CHV
 
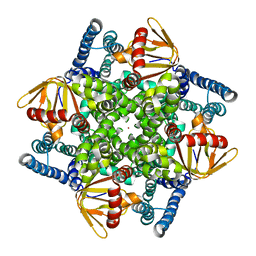 | | The electron crystallography structure of the cAMP-bound potassium channel MloK1 | | Descriptor: | CYCLIC NUCLEOTIDE-GATED POTASSIUM CHANNEL MLL3241, POTASSIUM ION | | Authors: | Kowal, J, Chami, M, Baumgartner, P, Arheit, M, Chiu, P.L, Rangl, M, Scheuring, S, Schroeder, G.F, Nimigean, C.M, Stahlberg, H. | | Deposit date: | 2013-12-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON CRYSTALLOGRAPHY (7 Å) | | Cite: | Ligand-Induced Structural Changes in the Cyclic Nucleotide-Modulated Potassium Channel Mlok1
Nat.Commun., 5, 2014
|
|
6I9D
 
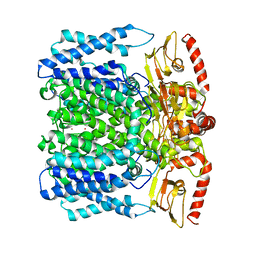 | | MloK1 consensus structure from single particle analysis of 2D crystals | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-11-23 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
6IAX
 
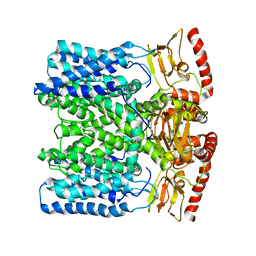 | | MloK1 model from single particle analysis of 2D crystals, class 1 (extended conformation) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Righetto, R, Biyani, N, Kowal, J, Chami, M, Stahlberg, H. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Retrieving high-resolution information from disordered 2D crystals by single-particle cryo-EM.
Nat Commun, 10, 2019
|
|
8J8Z
 
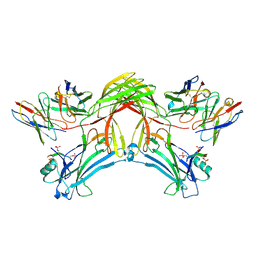 | | Structure of beta-arrestin1 in complex with D6Rpp | | Descriptor: | Atypical chemokine receptor 2, Beta-arrestin-1, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J9K
 
 | | Structure of basal beta-arrestin2 | | Descriptor: | Beta-arrestin-2, Fab6 heavy chain, Fab6 light chain | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8J8V
 
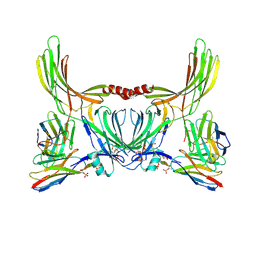 | | Structure of beta-arrestin2 in complex with D6Rpp (Local Refine) | | Descriptor: | Atypical chemokine receptor 2, Beta-arrestin-2, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-02 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
8JA3
 
 | | Structure of beta-arrestin1 in complex with C3aRpp | | Descriptor: | Beta-arrestin-1, C3a anaphylatoxin chemotactic receptor, Fab30 heavy chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Science, 383, 2024
|
|
6G2H
 
 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) core at 4.6 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
6G2I
 
 | | Filament of acetyl-CoA carboxylase and BRCT domains of BRCA1 (ACC-BRCT) at 5.9 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1, Breast cancer type 1 susceptibility protein | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
6G2D
 
 | | Citrate-induced acetyl-CoA carboxylase (ACC-Cit) filament at 5.4 A resolution | | Descriptor: | Acetyl-CoA carboxylase 1 | | Authors: | Hunkeler, M, Hagmann, A, Stuttfeld, E, Chami, M, Stahlberg, H, Maier, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Structural basis for regulation of human acetyl-CoA carboxylase.
Nature, 558, 2018
|
|
8IY9
 
 | | Structure of Niacin-GPR109A-G protein complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IYH
 
 | | Structure of MK6892-GPR109A-G-protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8IYW
 
 | | Structure of GSK256073-GPR109A-G-protein complex | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8JHN
 
 | | Structure of MMF-GPR109A-G protein complex | | Descriptor: | (E)-4-methoxy-4-oxidanylidene-but-2-enoic acid, G protein subunit alpha o1,Guanine nucleotide-binding protein G(o) subunit alpha, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-24 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8JER
 
 | | Structure of Acipimox-GPR109A-G protein complex | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yadav, M.K, Sarma, P, Chami, M, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-05-16 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-guided engineering of biased-agonism in the human niacin receptor via single amino acid substitution.
Nat Commun, 15, 2024
|
|
8RGZ
 
 | | Trimeric HSV-1F gB ectodomain in postfusion conformation with three bound HDIT101 Fab molecules. | | Descriptor: | Envelope glycoprotein B, HDIT101 Fab heavy chain, HDIT101 Fab light chain | | Authors: | Kalbermatter, D, Seyfizadeh, N, Imhof, T, Ries, M, Mueller, C, Jenner, L, Blumenschein, E, Yendrzheyevskiy, A, Moog, K, Eckert, D, Engel, R, Diebolder, P, Chami, M, Krauss, J, Schaller, T, Arndt, M. | | Deposit date: | 2023-12-14 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Development of a highly effective combination monoclonal antibody therapy against Herpes simplex virus.
J.Biomed.Sci., 31, 2024
|
|
