7M6S
 
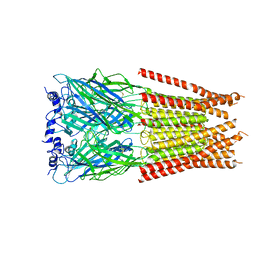 | |
7M6O
 
 | | Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
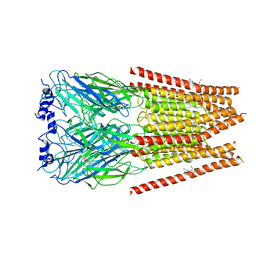
7M6P
 
 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7AV6
 
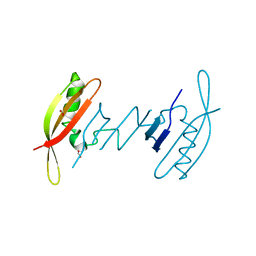 | | FAST in a domain-swapped dimer form | | Descriptor: | FORMIC ACID, Photoactive yellow protein | | Authors: | Bukhdruker, S, Remeeva, A, Ruchkin, D, Gorbachev, D, Povarova, N, Mineev, K, Goncharuk, S, Baranov, M, Mishin, A, Borshchevskiy, V. | | Deposit date: | 2020-11-04 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | NanoFAST: structure-based design of a small fluorogen-activating protein with only 98 amino acids.
Chem Sci, 12, 2021
|
|
4RUV
 
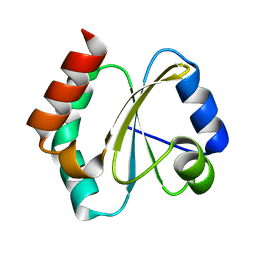 | | Crystal structure of thioredoxin 2 from Staphylococcus aureus NCTC8325 | | Descriptor: | Thioredoxin | | Authors: | Bose, M, Biswas, R, Roychowdhury, A, Bhattacharyya, S, Ghosh, A.K, Das, A.K. | | Deposit date: | 2014-11-22 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Elucidation of the mechanism of disulfide exchange between staphylococcal thioredoxin2 and thioredoxin reductase2: A structural insight.
Biochimie, 160, 2019
|
|
7D99
 
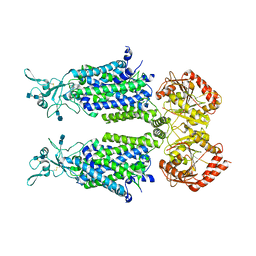 | | human potassium-chloride co-transporter KCC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
4RS3
 
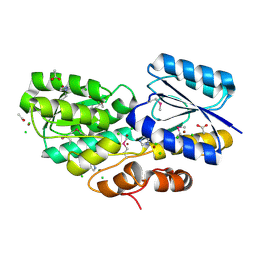 | | Crystal structure of carbohydrate transporter A0QYB3 from Mycobacterium smegmatis str. MC2 155, target EFI-510969, in complex with xylitol | | Descriptor: | ABC transporter, carbohydrate uptake transporter-2 (CUT2) family, periplasmic sugar-binding protein, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-06 | | Release date: | 2014-11-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
4RSM
 
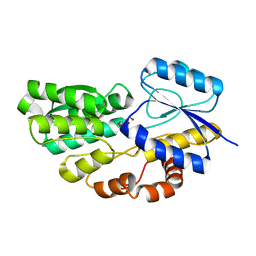 | | Crystal structure of carbohydrate transporter msmeg_3599 from mycobacterium smegmatis str. mc2 155, target efi-510970, in complex with d-threitol | | Descriptor: | D-Threitol, Periplasmic binding protein/LacI transcriptional regulator | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Hillerich, B, Siedel, R.D, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-08 | | Release date: | 2014-12-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A General Strategy for the Discovery of Metabolic Pathways: d-Threitol, l-Threitol, and Erythritol Utilization in Mycobacterium smegmatis.
J.Am.Chem.Soc., 137, 2015
|
|
7BV1
 
 | | Cryo-EM structure of the apo nsp12-nsp7-nsp8 complex | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
4UMC
 
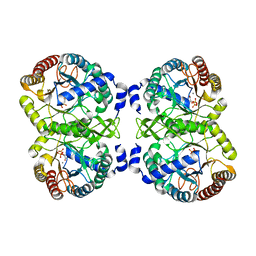 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | L-PHOSPHOLACTATE, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4UMB
 
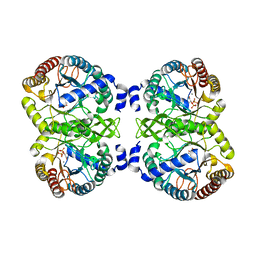 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase - the importance of accommodating the active site water | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
4UMA
 
 | | Structural analysis of substrate-mimicking inhibitors in complex with Neisseria meningitidis 3 deoxy D arabino heptulosonate 7 phosphate synthase the importance of accommodating the active site water | | Descriptor: | (E)-2-METHYL-3-PHOSPHONOACRYLATE, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE | | Authors: | Heyes, L.C, Reichau, S, Cross, P.J, Parker, E.J. | | Deposit date: | 2014-05-16 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Analysis of Substrate-Mimicking Inhibitors in Complex with Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase - the Importance of Accommodating the Active Site Water.
Bioorg.Chem., 57, 2014
|
|
8X3N
 
 | |
8X40
 
 | |
8XTH
 
 | |
6OY9
 
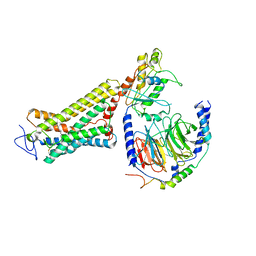 | | Structure of the Rhodopsin-Transducin Complex | | Descriptor: | Gt-alpha/Gi1-alpha chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, ... | | Authors: | Gao, Y, Hu, H, Ramachandran, S, Erickson, J.W, Cerione, R.A, Skiniotis, G. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2019-12-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the Rhodopsin-Transducin Complex: Insights into G-Protein Activation.
Mol.Cell, 75, 2019
|
|
7D90
 
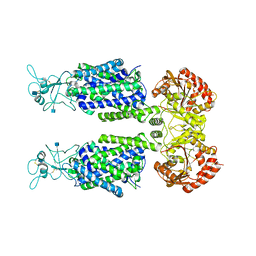 | | human potassium-chloride co-transporter KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D8Z
 
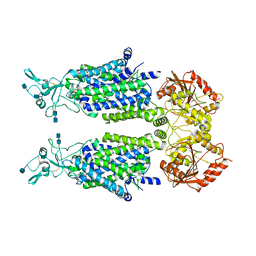 | | human potassium-chloride co-transporter KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
5J16
 
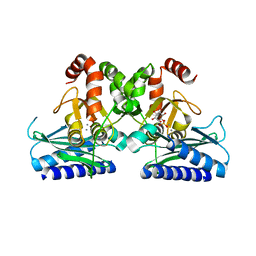 | |
6EZ8
 
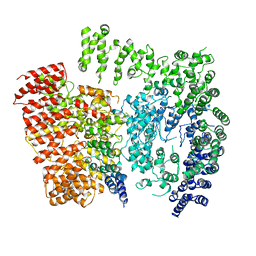 | | Human Huntingtin-HAP40 complex structure | | Descriptor: | Factor VIII intron 22 protein, Huntingtin | | Authors: | Guo, Q, Bin, H, Cheng, J, Pfeifer, G, Baumeister, W, Fernandez-Busnadiego, R, Kochanek, S. | | Deposit date: | 2017-11-14 | | Release date: | 2018-02-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-electron microscopy structure of huntingtin.
Nature, 555, 2018
|
|
7ACJ
 
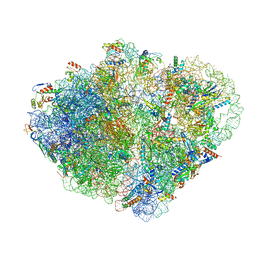 | | Structure of translocated trans-translation complex on E. coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
7ABZ
 
 | | Structure of pre-accomodated trans-translation complex on E. coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-09 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
7ACR
 
 | | Structure of post-translocated trans-translation complex on E. coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
7AC7
 
 | | Structure of accomodated trans-translation complex on E. Coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-10 | | Release date: | 2021-08-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
