7C0Z
 
 | | Crystal structure of a dinucleotide-binding protein (Y246A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form II) | | Descriptor: | 1,2-ETHANEDIOL, Sugar ABC transporter, periplasmic sugar-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C1B
 
 | | Crystal structure of a dinucleotide-binding protein (F79A/Y224A/Y246A and deletion of residues 50-75) of ABC transporter (unbound form) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sugar ABC transporter, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-02 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7C0W
 
 | | Crystal structure of a dinucleotide-binding protein (Y224A) of ABC transporter endogenously bound to uridylyl-3'-5'-phospho-guanosine (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, GLYCEROL, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-01 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and thermodynamic insights into the novel dinucleotide-binding protein of ABC transporter unveils its moonlighting function.
Febs J., 288, 2021
|
|
7LMT
 
 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a
To Be Published
|
|
6TH8
 
 | |
6BPI
 
 | | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates | | Descriptor: | 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, MLY-SER-THR-E2G, ... | | Authors: | MADER, P, Mendoza-Sanchez, R, DONG, A, DOBROVETSKY, E, IQBAL, A, CORLESS, V, TEMPEL, W, LIEW, S.K, SMIL, D, DELA SENA, C.C, KENNEDY, S, DIAZ, D.B, SCHAPIRA, M, VEDADI, M, BROWN, P.J, Santhakumar, V, FRYE, S, Bountra, C, Edwards, A.M, YUDIN, A.K, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain with aryl triazole fragment peptide conjugates
to be published
|
|
5ABW
 
 | | Neutrophil elastase inhibitors for the treatment of (cardio)pulmonary diseases | | Descriptor: | 1-(3-CHLOROPHENYL)-5-(3,5-DIMETHYLISOXAZOL-4-YL)-6-METHYL-N-[4-(METHYLSULFONYL)BENZYL]-2-OXO-1,2-DIHYDROPYRIDINE-3-CARBOXAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | von Nussbaum, F, Li, V.M, Schaefer, M. | | Deposit date: | 2015-08-10 | | Release date: | 2015-08-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Neutrophil elastase inhibitors for the treatment of (cardio)pulmonary diseases: Into clinical testing with pre-adaptive pharmacophores.
Bioorg. Med. Chem. Lett., 25, 2015
|
|
6TY3
 
 | | FAK structure from single particle analysis of 2D crystals | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Acebron, I, Righetto, R, Biyani, N, Chami, M, Boskovic, J, Stahlberg, H, Lietha, D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.32 Å) | | Cite: | Structural basis of Focal Adhesion Kinase activation on lipid membranes.
Embo J., 39, 2020
|
|
6TY4
 
 | | FAK structure with AMP-PNP from single particle analysis of 2D crystals | | Descriptor: | Focal adhesion kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Acebron, I, Righetto, R, Biyani, N, Chami, M, Boskovic, J, Stahlberg, H, Lietha, D. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.96 Å) | | Cite: | Structural basis of Focal Adhesion Kinase activation on lipid membranes.
Embo J., 39, 2020
|
|
7M40
 
 | | Discovery of small molecule antagonists of human Retinoblastoma Binding Protein 4 (RBBP4) | | Descriptor: | Histone-binding protein RBBP4, N~3~-{4-[3-(dimethylamino)pyrrolidin-1-yl]-6,7-dimethoxyquinazolin-2-yl}-N~1~,N~1~-dimethylpropane-1,3-diamine | | Authors: | Perveen, S, Dong, A, Tempel, W, Zepeda-Velazquez, C, Abbey, M, McLeod, D, Marcellus, R, Mohammed, M, Ensan, D, Panagopoulos, D, Trush, V, Gibson, E, Brown, P.J, Arrowsmith, C.H, Schapira, M, Al-awar, R, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of small molecule antagonists of human Retinoblastoma Binding Protein 4 (RBBP4)
To Be Published
|
|
6BP6
 
 | |
7MS7
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (5-((4-(4-chlorophenyl)piperidin-1-yl)sulfonyl)picolinoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-{5-[4-(4-chlorophenyl)piperidine-1-sulfonyl]pyridine-2-carbonyl}glycine, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS5
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-(4-(3,4-difluoro-phenyl)-piperidin-1-ylsulfonyl)-phenyl)-4-oxo-butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[4-(3,4-difluorophenyl)piperidine-1-sulfonyl]phenyl}-4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS6
 
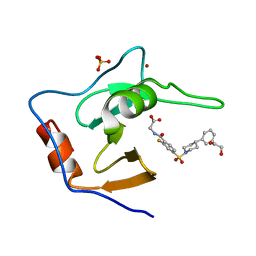 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (2-fluoro-4-((4-phenylpiperidin-1-yl)sulfonyl)benzoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, N-[2-fluoro-4-(4-phenylpiperidine-1-sulfonyl)benzoyl]glycine, SULFATE ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
4G2C
 
 | | DyP2 from Amycolatopsis sp. ATCC 39116 | | Descriptor: | ACETATE ION, DyP2, MANGANESE (II) ION, ... | | Authors: | Brown, M.E, Barros, T, Chang, M.C.Y. | | Deposit date: | 2012-07-11 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium.
Acs Chem.Biol., 7, 2012
|
|
7XCL
 
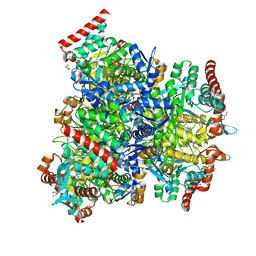 | |
7XCM
 
 | | Crystal structure of sulfite MttB structure at 3.2 A resolution | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
3KPD
 
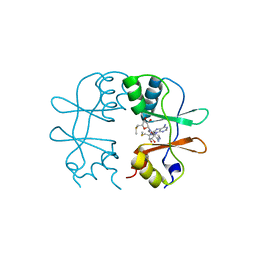 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine. | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | Authors: | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Chantar, M.L, Martinez-Cruz, L.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
7LGK
 
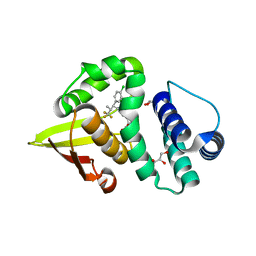 | | Crystal structure of soluble guanylate cyclase activator runcaciguat (BAY 1101042) bound to nostoc H-NOX domain | | Descriptor: | CHLORIDE ION, GLYCEROL, H-NOX domain protein, ... | | Authors: | van den Akker, F, Kumar, V, Schaefer, M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the Soluble Guanylate Cyclase Activator Runcaciguat (BAY 1101042).
J.Med.Chem., 64, 2021
|
|
3KPC
 
 | | Crystal Structure of the CBS domain pair of protein MJ0100 in complex with 5 -methylthioadenosine and S-adenosyl-L-methionine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, S-ADENOSYLMETHIONINE, Uncharacterized protein MJ0100 | | Authors: | Lucas, M, Oyenarte, I, Garcia, I.G, Arribas, E.A, Encinar, J.A, Kortazar, D, Fernandez, J.A, Mato, J.M, Martinez-Chantar, M.L, Martinez-Cruz, L.A. | | Deposit date: | 2009-11-16 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Binding of S-Methyl-5'-Thioadenosine and S-Adenosyl-l-Methionine to Protein MJ0100 Triggers an Open-to-Closed Conformational Change in Its CBS Motif Pair.
J.Mol.Biol., 396, 2010
|
|
4GGO
 
 | |
1MDX
 
 | | Crystal structure of ArnB transferase with pyridoxal 5' phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, ArnB aminotransferase, GLYCEROL, ... | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T.A, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
5FYQ
 
 | | Sirt2 in complex with a 13-mer trifluoroacetylated Ran peptide | | Descriptor: | NAD-DEPENDENT PROTEIN DEACETYLASE SIRTUIN-2, RAN AA 31-43, SULFATE ION, ... | | Authors: | Knyphausen, P, de Boor, S, Scislowski, L, Extra, A, Baldus, L, Schacherl, M, Baumann, U, Neundorf, I, Lammers, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights Into Lysine-Deacetylation of Natively Folded Substrate Proteins by Sirtuins.
J.Biol.Chem., 291, 2016
|
|
6BJ9
 
 | |
6BJB
 
 | |
