3MG8
 
 | | Structure of yeast 20S open-gate proteasome with Compound 16 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-(2,2-dimethylpropyl)-N~2~-[1H-indol-3-yl(oxo)acetyl]-L-asparaginyl-N-(2-methylbenzyl)-3-pyridin-4-yl-L-alaninamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
3MG4
 
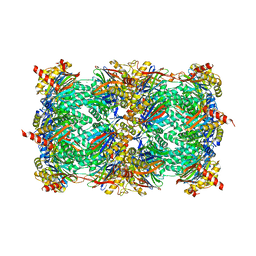 | | Structure of yeast 20S proteasome with Compound 1 | | Descriptor: | (2S)-2-amino-N-[(1S)-1-({(1S)-1-[(4-methylbenzyl)carbamoyl]-3-phenylpropyl}carbamoyl)-3-phenylpropyl]-4-phenylbutanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-04-05 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Characterization of a new series of non-covalent proteasome inhibitors with exquisite potency and selectivity for the 20S beta5-subunit.
Biochem.J., 430, 2010
|
|
1JR1
 
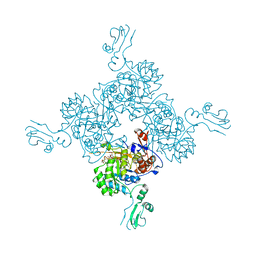 | | Crystal structure of Inosine Monophosphate Dehydrogenase in complex with Mycophenolic Acid | | Descriptor: | INOSINIC ACID, Inosine-5'-Monophosphate Dehydrogenase 2, MYCOPHENOLIC ACID, ... | | Authors: | Sintchak, M.D, Fleming, M.A, Futer, O, Raybuck, S.A, Chambers, S.P, Caron, P.R, Murcko, M.A, Wilson, K.P. | | Deposit date: | 2001-08-09 | | Release date: | 2001-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid.
Cell(Cambridge,Mass.), 85, 1996
|
|
3O75
 
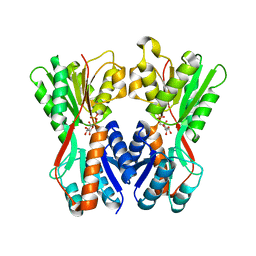 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida in complex with fructose 1-phosphate' | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, Fructose transport system repressor FruR | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
3OEU
 
 | | Structure of yeast 20S open-gate proteasome with Compound 24 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-{(2S)-1-[(2-chlorobenzyl)amino]-1-oxo-4-phenylbutan-2-yl}-N~2~-[3-(2-methylphenyl)propanoyl]-L-threoninamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2010-08-13 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4HUA
 
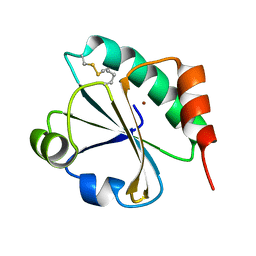 | | E. coli thioredoxin variant with (4R)-FluoroPro76 as single proline residue | | Descriptor: | COPPER (II) ION, Thioredoxin-1 | | Authors: | Scharer, M.A, Rubini, M, Capitani, G, Glockshuber, R. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | (4R)- and (4S)-Fluoroproline in the Conserved cis-Prolyl Peptide Bond of the Thioredoxin Fold: Tertiary Structure Context Dictates Ring Puckering.
Chembiochem, 14, 2013
|
|
3SDI
 
 | | Structure of yeast 20S open-gate proteasome with Compound 20 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N~4~-(2,2-dimethylpropyl)-N~1~-{(2S)-1-[(4-methylbenzyl)amino]-1-oxo-4-phenylbutan-2-yl}-N~2~-[(5-methyl-1,2-oxazol-3-yl)carbonyl]-L-aspartamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-06-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4HU9
 
 | | E. coli thioredoxin variant with (4S)-FluoroPro76 as single proline residue | | Descriptor: | COPPER (II) ION, Thioredoxin-1 | | Authors: | Scharer, M.A, Rubini, M, Capitani, G, Glockshuber, R. | | Deposit date: | 2012-11-02 | | Release date: | 2013-05-29 | | Last modified: | 2017-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | (4R)- and (4S)-Fluoroproline in the Conserved cis-Prolyl Peptide Bond of the Thioredoxin Fold: Tertiary Structure Context Dictates Ring Puckering.
Chembiochem, 14, 2013
|
|
1M1L
 
 | | Human Suppressor of Fused (N-terminal domain) | | Descriptor: | Suppressor of Fused | | Authors: | Merchant, M, Vajdos, F.F, Ultsch, M, Maun, H.R, Wendt, U, Cannon, J, Lazarus, R.A, de Vos, A.M, de Sauvage, F.J. | | Deposit date: | 2002-06-19 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Suppressor of fused regulates Gli activity through a dual binding mechanism
Mol.Cell.Biol., 24, 2004
|
|
1A7Y
 
 | | CRYSTAL STRUCTURE OF ACTINOMYCIN D | | Descriptor: | ACTINOMYCIN D, ETHYL ACETATE, METHANOL | | Authors: | Schafer, M, Sheldrick, G.M, Bahner, I, Lackner, H. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
1A7Z
 
 | | CRYSTAL STRUCTURE OF ACTINOMYCIN Z3 | | Descriptor: | ACTINOMYCIN Z3, BENZENE | | Authors: | Schafer, M. | | Deposit date: | 1998-03-19 | | Release date: | 1999-03-23 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal Structures of Actinomycin D and Actinomycin Z3.
Angew.Chem.Int.Ed.Engl., 37, 1998
|
|
4G4I
 
 | | Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile determined at 1.9 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-glucuronoyl methylesterase, GLYCEROL | | Authors: | Charvagi, M.D, Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2012-07-16 | | Release date: | 2013-01-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3SQB
 
 | | Structure of the major type 1 pilus subunit FimA bound to the FimC chaperone | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chaperone protein fimC, ... | | Authors: | Scharer, M.A, Eidam, O, Grutter, M.G, Glockshuber, R, Capitani, G. | | Deposit date: | 2011-07-05 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
4BGB
 
 | |
4BG9
 
 | | Apo form of a putative sugar kinase MK0840 from Methanopyrus kandleri (orthorhombic space group) | | Descriptor: | ACETATE ION, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Schacherl, M, Waltersperger, S.M, Baumann, U. | | Deposit date: | 2013-03-24 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Characterization of the Ribonuclease H-Like Type Askha Superfamily Kinase Mk0840 from Methanopyrus Kandleri
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4NET
 
 | | Crystal structure of ADC-1 beta-lactamase | | Descriptor: | AmpC, GLYCEROL, NITRATE ION | | Authors: | Bhattacharya, M, Toth, M, Antunes, N.T, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of the extended-spectrum class C beta-lactamase ADC-1 from Acinetobacter baumannii.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3Q96
 
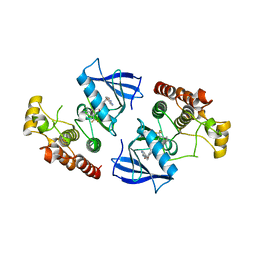 | | B-Raf kinase domain in complex with a tetrahydronaphthalene inhibitor | | Descriptor: | (2S)-N-[3-(2-aminopropan-2-yl)-5-(trifluoromethyl)phenyl]-7-[(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-4-yl)oxy]-1,2,3,4-tetrahydronaphthalene-2-carboxamide, Serine/threonine-protein kinase B-raf | | Authors: | Sintchak, M.D, Aertgeerts, K, Yano, J. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Design and optimization of potent and orally bioavailable tetrahydronaphthalene raf inhibitors.
J.Med.Chem., 54, 2011
|
|
4DPV
 
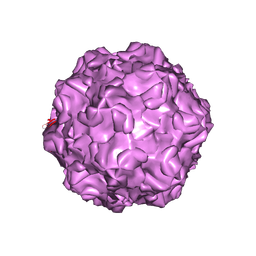 | | PARVOVIRUS/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*CP*TP*CP*TP*TP*GP*C)-3'), MAGNESIUM ION, PROTEIN (PARVOVIRUS COAT PROTEIN) | | Authors: | Chapman, M.S, Rossmann, M.G. | | Deposit date: | 1996-02-01 | | Release date: | 1997-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Canine parvovirus capsid structure, analyzed at 2.9 A resolution.
J.Mol.Biol., 264, 1996
|
|
3SDK
 
 | | Structure of yeast 20S open-gate proteasome with Compound 34 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, N-[(2S)-3-(3-tert-butyl-1,2,4-oxadiazol-5-yl)-1-({(2S)-1-[(4-methylbenzyl)amino]-1-oxo-4-phenylbutan-2-yl}amino)-1-oxopropan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2011-06-09 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Optimization of a series of dipeptides with a P3 threonine residue as non-covalent inhibitors of the chymotrypsin-like activity of the human 20S proteasome.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4DWH
 
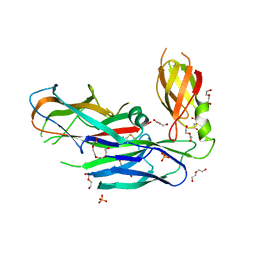 | | Structure of the major type 1 pilus subunit FIMA bound to the FIMC (2.5 A resolution) | | Descriptor: | Chaperone protein fimC, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Scharer, M.A, Puorger, C, Crespo, M, Glockshuber, R, Capitani, G. | | Deposit date: | 2012-02-24 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
3PIU
 
 | |
4G4J
 
 | | Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile in complex with methyl 4-O-methyl-beta-D-glucopyranuronate determined at 2.35 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-glucuronoyl methylesterase, GLYCEROL, ... | | Authors: | Charavgi, M.D, Dimarogona, M, Topakas, E, Christakopoulos, P, Chrysina, E.D. | | Deposit date: | 2012-07-16 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The structure of a novel glucuronoyl esterase from Myceliophthora thermophila gives new insights into its role as a potential biocatalyst.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4MCK
 
 | |
4BGA
 
 | |
1L1L
 
 | | CRYSTAL STRUCTURE OF B-12 DEPENDENT (CLASS II) RIBONUCLEOTIDE REDUCTASE | | Descriptor: | RIBONUCLEOSIDE TRIPHOSPHATE REDUCTASE | | Authors: | Sintchak, M.D, Arjara, G, Kellogg, B.A, Stubbe, J, Drennan, C.L. | | Deposit date: | 2002-02-18 | | Release date: | 2002-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of class II ribonucleotide reductase reveals how an allosterically regulated monomer mimics a dimer.
Nat.Struct.Biol., 9, 2002
|
|
