6P74
 
 | | OLD nuclease from Thermus Scotoductus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PLATINUM (II) ION, Putative ATP-dependent endonuclease of the OLD family, ... | | Authors: | Chappie, J.S, Schiltz, C.J. | | Deposit date: | 2019-06-04 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The full-length structure of Thermus scotoductus OLD defines the ATP hydrolysis properties and catalytic mechanism of Class 1 OLD family nucleases.
Nucleic Acids Res., 48, 2020
|
|
3ZYS
 
 | | Human dynamin 1 deltaPRD polymer stabilized with GMPPCP | | Descriptor: | DYNAMIN-1, INTERFERON-INDUCED GTP-BINDING PROTEIN MX1 | | Authors: | Chappie, J.S, Mears, J.A, Fang, S, Leonard, M, Schmid, S.L, Milligan, R.A, Hinshaw, J.E, Dyda, F. | | Deposit date: | 2011-08-24 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.2 Å) | | Cite: | A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke.
Cell(Cambridge,Mass.), 147, 2011
|
|
2X2E
 
 | | Dynamin GTPase dimer, long axis form | | Descriptor: | DYNAMIN-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chappie, J.S, Acharya, S, Leonard, M, Schmid, S.L, Dyda, F. | | Deposit date: | 2010-01-12 | | Release date: | 2010-04-28 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | G Domain Dimerization Controls Dynamin'S Assembly-Stimulated Gtpase Activity.
Nature, 465, 2010
|
|
2X2F
 
 | | Dynamin 1 GTPase dimer, short axis form | | Descriptor: | DYNAMIN-1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chappie, J.S, Acharya, S, Leonard, M, Schmid, S.L, Dyda, F. | | Deposit date: | 2010-01-13 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | G Domain Dimerization Controls Dynamin'S Assembly-Stimulated Gtpase Activity.
Nature, 465, 2010
|
|
3ZYC
 
 | | DYNAMIN 1 GTPASE GED FUSION DIMER COMPLEXED WITH GMPPCP | | Descriptor: | DYNAMIN-1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Chappie, J.S, Mears, J.A, Fang, S, Leonard, M, Schmid, S.L, Milligan, R.A, Hinshaw, J.E, Dyda, F. | | Deposit date: | 2011-08-22 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Pseudoatomic Model of the Dynamin Polymer Identifies a Hydrolysis-Dependent Powerstroke.
Cell(Cambridge,Mass.), 147, 2011
|
|
6KF4
 
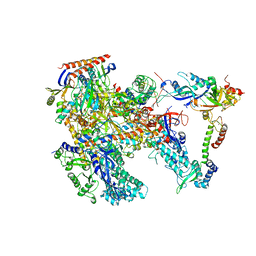 | | Cryo-EM structure of Thermococcus kodakarensis RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit A'', DNA-directed RNA polymerase subunit D, ... | | Authors: | Jun, S.-H, Hyun, J, Jeong, H, Cha, J.S, Kim, H, Bartlett, M.S, Cho, H.-S, Murakami, K.S. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-01 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Direct binding of TFE alpha opens DNA binding cleft of RNA polymerase.
Nat Commun, 11, 2020
|
|
6LBX
 
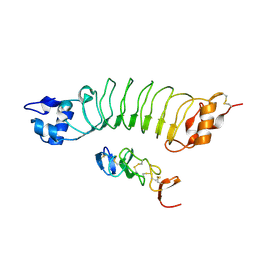 | | Crystal structure of HER2 Domain IV and Rb-H2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-2, Repebody (Rb-H2) | | Authors: | Cho, H.S, Cha, J.S. | | Deposit date: | 2019-11-15 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Computationally-guided design and affinity improvement of a protein binder targeting a specific site on HER2
Comput Struct Biotechnol J, 19, 2021
|
|
4V9D
 
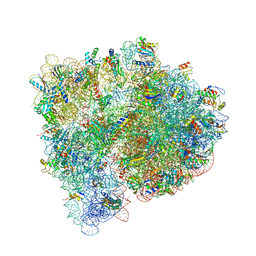 | | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Wang, L, Feldman, M.B, Pulk, A, Chen, V.B, Kapral, G.J, Noeske, J, Richardson, J.S, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2012-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding.
Science, 332, 2011
|
|
1VJK
 
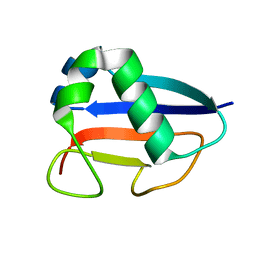 | | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 | | Descriptor: | molybdopterin converting factor, subunit 1 | | Authors: | Chen, L, Liu, Z.J, Tempel, W, Shah, A, Lee, D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 '
To be published
|
|
1VK1
 
 | | Conserved hypothetical protein from Pyrococcus furiosus Pfu-392566-001 | | Descriptor: | Conserved hypothetical protein, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Shah, A, Liu, Z.J, Tempel, W, Chen, L, Lee, D, Yang, H, Chang, J, Zhao, M, Ng, J, Rose, J, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | (NZ)CH...O contacts assist crystallization of a ParB-like nuclease.
Bmc Struct.Biol., 7, 2007
|
|
3C48
 
 | |
3C4V
 
 | | Structure of the retaining glycosyltransferase MshA:The first step in mycothiol biosynthesis. Organism: Corynebacterium glutamicum : Complex with UDP and 1L-INS-1-P. | | Descriptor: | L-MYO-INOSITOL-1-PHOSPHATE, MAGNESIUM ION, Predicted glycosyltransferases, ... | | Authors: | Vetting, M.W, Frantom, P.A, Blanchard, J.S. | | Deposit date: | 2008-01-30 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Enzymatic Analysis of MshA from Corynebacterium glutamicum: SUBSTRATE-ASSISTED CATALYSIS
J.Biol.Chem., 283, 2008
|
|
3C8Z
 
 | | The 1.6 A Crystal Structure of MshC: The Rate Limiting Enzyme in the Mycothiol Biosynthetic Pathway | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, Cysteinyl-tRNA synthetase, ... | | Authors: | Tremblay, L.W, Fan, F, Vetting, M.W, Blanchard, J.S. | | Deposit date: | 2008-02-14 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of Mycobacterium smegmatis MshC: the penultimate enzyme in the mycothiol biosynthetic pathway.
Biochemistry, 47, 2008
|
|
3CG5
 
 | |
4E3R
 
 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4E3Q
 
 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
1M44
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-APO Structure | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, SULFATE ION | | Authors: | Vetting, M.W, Hegde, S.S, Javid-Majd, F, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
Nat.Struct.Biol., 9, 2002
|
|
1M4D
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Tobramycin | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside 2'-N-acetyltransferase, COENZYME A, ... | | Authors: | Vetting, M.W, Hegde, S.S, Javid-Majd, F, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
Nat.Struct.Biol., 9, 2002
|
|
1M4I
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Kanamycin A | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside 2'-N-acetyltransferase, COENZYME A, ... | | Authors: | Vetting, M.W, Hegde, S.S, Javid-Majd, F, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
Nat.Struct.Biol., 9, 2002
|
|
1M4G
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-Complex with Coenzyme A and Ribostamycin | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, Aminoglycoside 2'-N-acetyltransferase, COENZYME A, ... | | Authors: | Vetting, M.W, Hegde, S.S, Javid-Majd, F, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
Nat.Struct.Biol., 9, 2002
|
|
9CSY
 
 | | SARS-CoV-2 papain-like protease (PLpro) bound to PF-07957472 | | Descriptor: | 2-methyl-5-(4-methylpiperazin-1-yl)-N-{1-[(2P)-2-(1-methyl-1H-pyrazol-4-yl)quinolin-4-yl]cyclopropyl}benzamide, Papain-like protease, ZINC ION, ... | | Authors: | Mashalidis, E.H, Chang, J.S, Wu, H, Garnsey, M. | | Deposit date: | 2024-07-24 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Discovery of SARS-CoV-2 papain-like protease (PL pro ) inhibitors with efficacy in a murine infection model.
Sci Adv, 10, 2024
|
|
4UUD
 
 | | Human dynamin 1 K44A superconstricted polymer stabilized with GTP | | Descriptor: | DYNAMIN-1 | | Authors: | Sundborger, A.C, Fang, S, Heymann, J.A, Ray, P, Chappie, J.S, Hinshaw, J.E. | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | A Dynamin Mutant Defines a Superconstricted Prefission State.
Cell Rep., 8, 2014
|
|
8TWQ
 
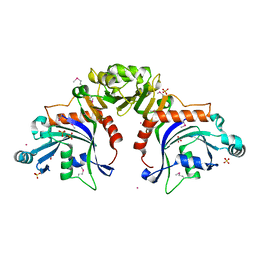 | | Structure of bacteriophage lambda RexA protein | | Descriptor: | CADMIUM ION, Protein rexA, SULFATE ION | | Authors: | Adams, M.C, Chappie, J.S, Schiltz, C.J. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of bacteriophage lambda RexA provides novel insights into the DNA binding properties of Rex-like phage exclusion proteins.
Nucleic Acids Res., 52, 2024
|
|
6UT6
 
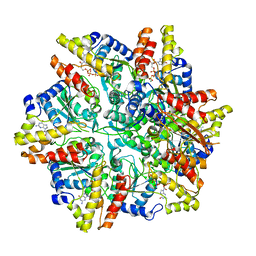 | | Cryo-EM structure of the Escherichia coli McrBC complex | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, 5-methylcytosine-specific restriction enzyme B, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Niu, Y, Suzuki, H, Hosford, C.J, Chappie, J.S, Walz, T. | | Deposit date: | 2019-10-29 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural asymmetry governs the assembly and GTPase activity of McrBC restriction complexes.
Nat Commun, 11, 2020
|
|
6UT3
 
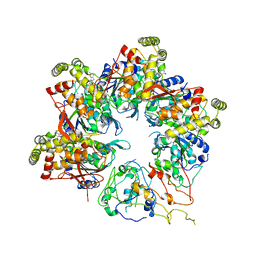 | |
