7PGT
 
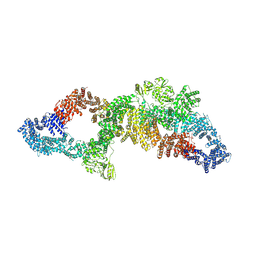 | | The structure of human neurofibromin isoform 2 in opened conformation. | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGS
 
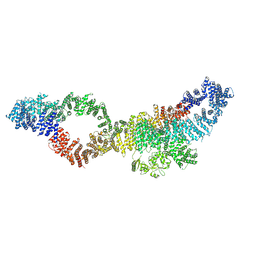 | | Consensus structure of human Neurofibromin isoform 2 | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGR
 
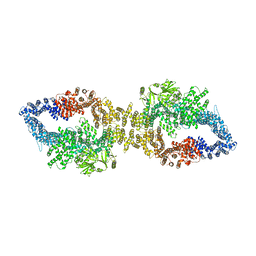 | | The structure of human neurofibromin isoform 2 in closed conformation | | Descriptor: | Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGU
 
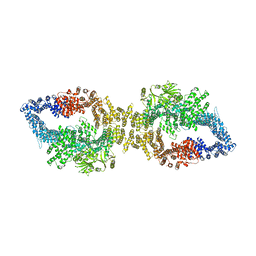 | | Autoinhibited structure of human neurofibromin isoform 2 stabilized by Zinc. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
7PGP
 
 | |
7PGQ
 
 | | GAP-SecPH region of human neurofibromin isoform 2 in closed conformation. | | Descriptor: | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Neurofibromin, ZINC ION | | Authors: | Naschberger, A, Baradaran, R, Carroni, M, Rupp, B. | | Deposit date: | 2021-08-15 | | Release date: | 2022-10-26 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The structure of neurofibromin isoform 2 reveals different functional states.
Nature, 599, 2021
|
|
6T8H
 
 | | Cryo-EM structure of the DNA-bound PolD-PCNA processive complex from P. abyssi | | Descriptor: | DNA polymerase II small subunit, DNA polymerase sliding clamp, DNA primer, ... | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Pehau-Arnaudet, G, England, P, Lindhal, E, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-24 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-08 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
5AFB
 
 | | Crystal structure of the Latrophilin3 Lectin and Olfactomedin Domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Jackson, V.A, del Toro, D, Carrasquero, M, Roversi, P, Harlos, K, Klein, R, Seiradake, E. | | Deposit date: | 2015-01-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis of Latrophilin-Flrt Interaction.
Structure, 23, 2015
|
|
6T7Y
 
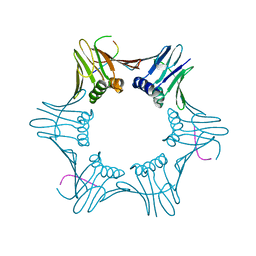 | | Structure of PCNA bound to cPIP motif of DP2 from P. abyssi | | Descriptor: | DNA polymerase sliding clamp, cPIP motif from the DP2 large subunit of PolD | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
6T7X
 
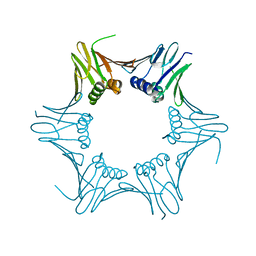 | | Crystal structure of PCNA from P. abyssi | | Descriptor: | DNA polymerase sliding clamp | | Authors: | Madru, C, Raia, P, Hugonneau Beaufet, I, Delarue, M, Carroni, M, Sauguet, L. | | Deposit date: | 2019-10-23 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA.
Nat Commun, 11, 2020
|
|
1ZUJ
 
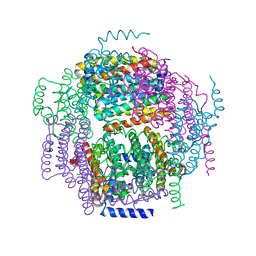 | | The crystal structure of the Lactococcus lactis MG1363 DpsA protein | | Descriptor: | hypothetical protein Llacc01001955 | | Authors: | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | Deposit date: | 2005-05-31 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
1ZS3
 
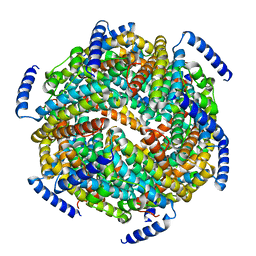 | | The crystal structure of the Lactococcus lactis MG1363 DpsB protein | | Descriptor: | Lactococcus lactis MG1363 DpsA | | Authors: | Stillman, T.J, Upadhyay, M, Norte, V.A, Sedelnikova, S.E, Carradus, M, Tzokov, S, Bullough, P.A, Shearman, C.A, Gasson, M.J, Williams, C.H, Artymiuk, P.J, Green, J. | | Deposit date: | 2005-05-23 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding.
Mol.Microbiol., 57, 2005
|
|
6FU8
 
 | |
6FB3
 
 | | Teneurin 2 Partial Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Teneurin-2, ... | | Authors: | Jackson, V.A, Carrasquero, M, Lowe, E.D, Seiradake, E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-03-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structures of Teneurin adhesion receptors reveal an ancient fold for cell-cell interaction.
Nat Commun, 9, 2018
|
|
1XU6
 
 | | Structure of the C-terminal domain from Trypanosoma brucei Variant Surface Glycoprotein MITat1.2 | | Descriptor: | Variant surface glycoprotein MITAT 1.2 | | Authors: | Chattopadhyay, A, Jones, N.G, Nietlispach, D, Nielsen, P.R, Voorheis, H.P, Mott, H.R, Carrington, M. | | Deposit date: | 2004-10-25 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-terminal domain from Trypanosoma brucei variant surface glycoprotein MITat1.2
J.Biol.Chem., 280, 2004
|
|
6SOY
 
 | | Trypanosoma brucei transferrin receptor in complex with human transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ESAG6, subunit of heterodimeric transferrin receptor, ... | | Authors: | Trevor, C, Carrington, M, Higgins, M.K. | | Deposit date: | 2019-08-30 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of the trypanosome transferrin receptor reveals mechanisms of ligand recognition and immune evasion.
Nat Microbiol, 4, 2019
|
|
6SOZ
 
 | | Glycosylated Trypanosoma brucei transferrin receptor in complex with human transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ESAG6, ... | | Authors: | Trevor, C, Carrington, M, Higgins, M.K. | | Deposit date: | 2019-08-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structure of the trypanosome transferrin receptor reveals mechanisms of ligand recognition and immune evasion.
Nat Microbiol, 4, 2019
|
|
6HMS
 
 | | Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3'), DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3'), DNA polymerase II large subunit,DNA polymerase II large subunit, ... | | Authors: | Raia, P, Carroni, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
3TTI
 
 | | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 10, trans-4-({9-[(3S)-tetrahydrofuran-3-yl]-8-[(2,4,6-trifluorophenyl)amino]-9H-purin-2-yl}amino)cyclohexanol | | Authors: | Plantevin-Krenitsky, V, Nadolny, L, Delgado, M, Ayala, L, Clareen, S, Hilgraf, R, Albers, R, Hegde, S, D'Sidocky, N, Sapienza, J, Wright, J, McCarrick, M, Bahmanyar, S, Chamberlain, P, Delker, S.L, Muir, J, Giegel, D, Xu, L, Celeridad, M, Lachowitzer, J, Bennett, B, Moghaddam, M, Khatsenko, O, Katz, J, Fan, R, Bai, A, Tang, Y, Shirley, M.A, Benish, B, Bodine, T, Blease, K, Raymon, H, Cathers, B.E, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
8PYQ
 
 | | 10 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
8PYO
 
 | | 5 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
8PYP
 
 | | 25 micrometer HEWL crystals solved at room-temperature using fixed-target serial crystallography. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Mason, T.J, Carrillo, M, Beale, J.H, Padeste, C. | | Deposit date: | 2023-07-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Micro-structured polymer fixed targets for serial crystallography at synchrotrons and XFELs.
Iucrj, 10, 2023
|
|
2JWH
 
 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
2JWG
 
 | | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein | | Descriptor: | Variant surface glycoprotein ILTAT 1.24 | | Authors: | Jones, N.G, Nietlispach, D, Sharma, R, Burke, D.F, Eyres, I, Mues, M, Mott, H.R, Carrington, M. | | Deposit date: | 2007-10-12 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of a Glycosylphosphatidylinositol-anchored Domain from a Trypanosome Variant Surface Glycoprotein
J.Biol.Chem., 283, 2008
|
|
4E40
 
 | | The haptoglobin-hemoglobin receptor of Trypanosoma congolense | | Descriptor: | Putative uncharacterized protein | | Authors: | Higgins, M.K, Tkachenko, O, Brown, A, Reed, J, Carrington, M. | | Deposit date: | 2012-03-11 | | Release date: | 2013-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the trypanosome haptoglobin-hemoglobin receptor and implications for nutrient uptake and innate immunity.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
