4B76
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, [2,4-bis(fluoranyl)-3-phenoxy-phenyl]methylazanium | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
4B75
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-amino-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
4B6F
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (4-phenoxyphenyl)methylazanium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3, ... | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
4B73
 
 | | Discovery of an allosteric mechanism for the regulation of HCV NS3 protein function | | Descriptor: | (2S)-4-amino-N-[(1R)-1-(4-chloro-2-fluoro-3-phenoxyphenyl)propyl]-4-oxobutan-2-aminium, NON-STRUCTURAL PROTEIN 4A, SERINE PROTEASE NS3 | | Authors: | Saalau-Bethell, S.M, Woodhead, A.J, Chessari, G, Carr, M.G, Coyle, J, Graham, B, Hiscock, S.D, Murray, C.W, Pathuri, P, Rich, S.J, Richardson, C.J, Williams, P.A, Jhoti, H. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of an Allosteric Mechanism for the Regulation of Hcv Ns3 Protein Function.
Nat.Chem.Biol., 8, 2012
|
|
1GYO
 
 | | Crystal structure of the di-tetraheme cytochrome c3 from Desulfovibrio gigas at 1.2 Angstrom resolution | | Descriptor: | CYTOCHROME C3, A DIMERIC CLASS III C-TYPE CYTOCHROME, GLYCEROL, ... | | Authors: | Aragao, D, Frazao, C, Sieker, L, Sheldrick, G.M, Legall, J, Carrondo, M.A. | | Deposit date: | 2002-04-29 | | Release date: | 2002-05-24 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Dimeric Cytochrome C3 from Desulfovibrio Gigas at 1.2 A Resolution
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3KFY
 
 | | Dynamic switching and partial occupancies of a small molecule inhibitor complex of DHFR | | Descriptor: | 5-[(4-chlorophenyl)sulfanyl]quinazoline-2,4-diamine, CALCIUM ION, Dihydrofolate reductase, ... | | Authors: | Collins, E.J, Lee, A.L, Carroll, M.J, Gromova, A.V, Miller, K.R, Singleton, S.F. | | Deposit date: | 2009-10-28 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Dynamic switching and partial occupancies of a small molecule inhibitor complex of DHFR
To be Published
|
|
6QB8
 
 | | Human CCT:mLST8 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cuellar, J, Santiago, C, Ludlam, W.G, Bueno-Carrasco, M.T, Valpuesta, J.M, Willardson, B.M. | | Deposit date: | 2018-12-20 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural and functional analysis of the role of the chaperonin CCT in mTOR complex assembly.
Nat Commun, 10, 2019
|
|
6ZJK
 
 | |
6ZVP
 
 | | Atomic model of the EM-based structure of the full-length tyrosine hydroxylase in complex with dopamine (residues 40-497) in which the regulatory domain (residues 40-165) has been included only with the backbone atoms | | Descriptor: | FE (III) ION, L-DOPAMINE, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-07-27 | | Release date: | 2021-11-17 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6ZZU
 
 | | Partial structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-08-05 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7A2G
 
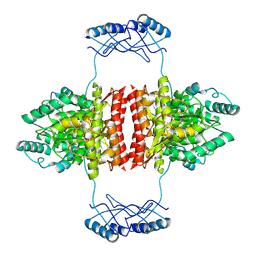 | | Full-length structure of the substrate-free tyrosine hydroxylase (apo-TH). | | Descriptor: | FE (III) ION, Tyrosine 3-monooxygenase | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Flydal, M.I, Martinez, A, Valpuesta, J.M. | | Deposit date: | 2020-08-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
6ZN2
 
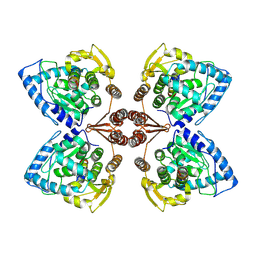 | | Partial structure of tyrosine hydroxylase in complex with dopamine showing the catalytic domain and an alpha-helix from the regulatory domain involved in dopamine binding. | | Descriptor: | FE (III) ION, L-DOPAMINE, SER-LEU-ILE-GLU-ASP-ALA-ARG-LYS-GLU-ARG-GLU-ALA-ALA-VAL-ALA-ALA-ALA-ALA, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2020-07-06 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7PIM
 
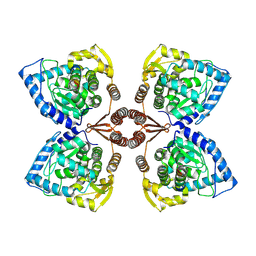 | | Partial structure of tyrosine hydroxylase lacking the first 35 residues in complex with dopamine. | | Descriptor: | FE (III) ION, L-DOPAMINE, Regulatory domain alpha-helix, ... | | Authors: | Bueno-Carrasco, M.T, Cuellar, J, Santiago, C, Valpuesta, J.M, Martinez, A, Flydal, M.I. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural mechanism for tyrosine hydroxylase inhibition by dopamine and reactivation by Ser40 phosphorylation.
Nat Commun, 13, 2022
|
|
7P37
 
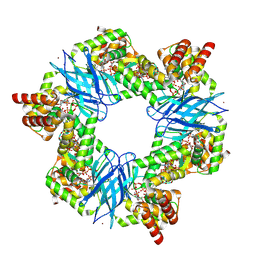 | | Streptomyces coelicolor ATP-loaded NrdR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ZINC ION | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P3Q
 
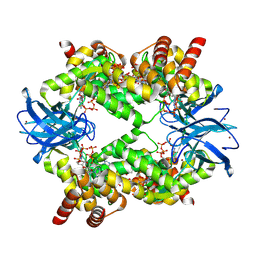 | | Streptomyces coelicolor dATP/ATP-loaded NrdR octamer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ... | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P3F
 
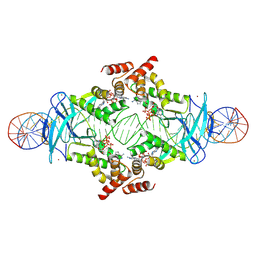 | |
6ZGK
 
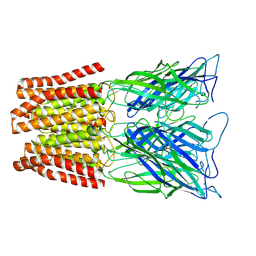 | | GLIC pentameric ligand-gated ion channel, pH 3 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6ZGD
 
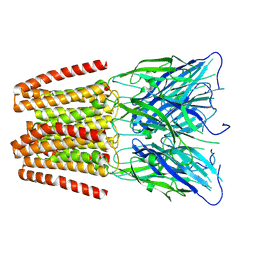 | | GLIC pentameric ligand-gated ion channel, pH 7 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6ZGJ
 
 | | GLIC pentameric ligand-gated ion channel, pH 5 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
8BYP
 
 | |
8C5Y
 
 | | RPA tetrameric supercomplex from Pyrococcus abyssi | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Legrand, P, Sauguet, L. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8C5Z
 
 | | RPA tetrameric supercomplex with AROD-OB-1 | | Descriptor: | RPA14 subunit of the hetero-oligomeric complex involved in homologous recombination, RPA32 subunit of the hetero-oligomeric complex involved in homologous recombination, Replication factor A, ... | | Authors: | Madru, C, Martinez-Carranza, M, Legrand, P, Sauguet, L. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | DNA-binding mechanism and evolution of replication protein A.
Nat Commun, 14, 2023
|
|
8C5H
 
 | |
7QFQ
 
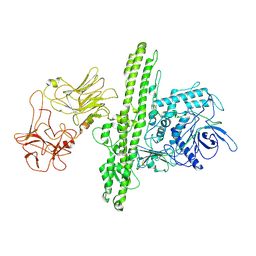 | | Cryo-EM structure of Botulinum neurotoxin serotype B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
7QFP
 
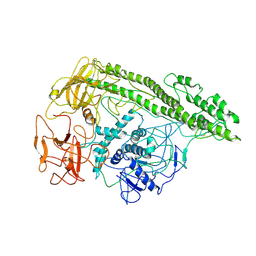 | | Cryo-EM structure of Botulinum neurotoxin serotype E | | Descriptor: | Botulinum neurotoxin | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
