2R8J
 
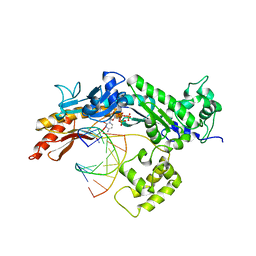 | | Structure of the Eukaryotic DNA Polymerase eta in complex with 1,2-d(GpG)-cisplatin containing DNA | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Cisplatin, ... | | Authors: | Carell, T, Alt, A, Lammens, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta
Science, 318, 2007
|
|
2R8K
 
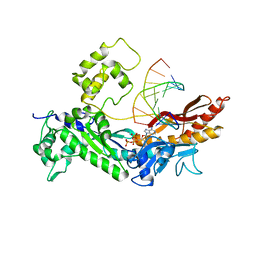 | | Structure of the Eukaryotic DNA Polymerase eta in complex with 1,2-d(GpG)-cisplatin containing DNA | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*DGP*DTP*DGP*DGP*DTP*DGP*DAP*DGP*DC)-3', 5'-D(P*DGP*DGP*DCP*DTP*DCP*DAP*DCP*DCP*DAP*DC)-3', ... | | Authors: | Carell, T, Alt, A, Lammens, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta
Science, 318, 2007
|
|
4UQG
 
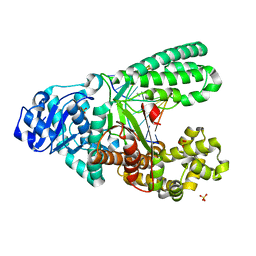 | | A new bio-isosteric base pair based on reversible bonding | | Descriptor: | 5'-D(*AP*GP*GP*GP*A SAYP*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*C T0TP*TP*CP*CP*CP*TP)-3', DNA POLYMERASE, ... | | Authors: | Tomas-Gamasa, M, Serdjukov, S, Su, M, Mueller, M, Carell, T. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | "Post-it" type connected DNA created with a reversible covalent cross-link.
Angew. Chem. Int. Ed. Engl., 54, 2015
|
|
6H0S
 
 | |
6FL1
 
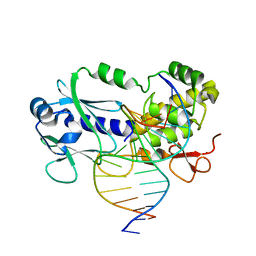 | | Crystal structure of the complex between the Lactococcus lactis FPG mutant T221P and a Fapy-dG containing DNA | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*TP*TP(FOX)P*TP*TP*TP*CP*TP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Castaing, B, Ober, M, Carell, T. | | Deposit date: | 2018-01-25 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the complex between the Lactococcus lactis FPG mutant T221P and a Fapy-dG containing DNA
To Be Published
|
|
4FHD
 
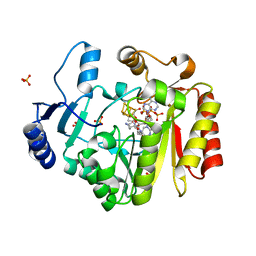 | | Spore photoproduct lyase complexed with dinucleoside spore photoproduct | | Descriptor: | 1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-[[(5R)-1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-methyl-2,4-bis(oxidanylidene)-1,3-diazinan-5-yl]methyl]pyrimidine-2,4-dione, IRON/SULFUR CLUSTER, PYROPHOSPHATE 2-, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
4FHG
 
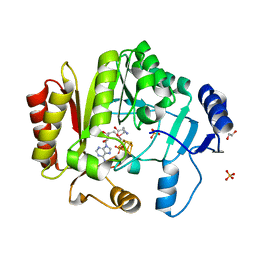 | | Spore photoproduct lyase C140S mutant | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
4FHE
 
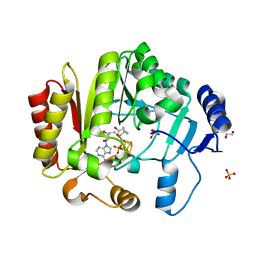 | | Spore photoproduct lyase C140A mutant | | Descriptor: | 1,2-ETHANEDIOL, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
4FHF
 
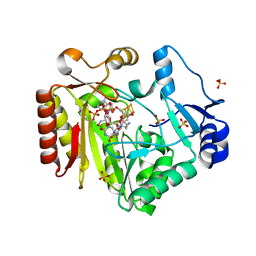 | | Spore photoproduct lyase C140A mutant with dinucleoside spore photoproduct | | Descriptor: | 1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-[[(5R)-1-[(2R,4S,5R)-5-(hydroxymethyl)-4-oxidanyl-oxolan-2-yl]-5-methyl-2,4-bis(oxidanylidene)-1,3-diazinan-5-yl]methyl]pyrimidine-2,4-dione, IRON/SULFUR CLUSTER, PYROPHOSPHATE 2-, ... | | Authors: | Benjdia, A, Heil, K, Barends, T.R.M, Carell, T, Schlichting, I. | | Deposit date: | 2012-06-06 | | Release date: | 2012-07-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into recognition and repair of UV-DNA damage by Spore Photoproduct Lyase, a radical SAM enzyme.
Nucleic Acids Res., 40, 2012
|
|
1XC8
 
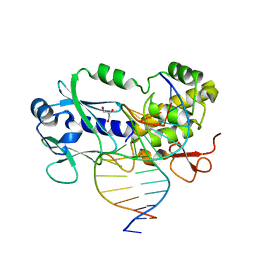 | | CRYSTAL STRUCTURE COMPLEX BETWEEN THE WILD-TYPE LACTOCOCCUS LACTIS FPG (MUTM) AND A FAPY-DG CONTAINING DNA | | Descriptor: | 5'-D(*CP*TP*CP*TP*TP*TP*(FOX)P*TP*TP*TP*CP*TP*CP*G)-3', 5'-D(*GP*CP*GP*AP*GP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Coste, F, Ober, M, Carell, T, Boiteux, S, Zelwer, C, Castaing, B. | | Deposit date: | 2004-09-01 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase.
J.Biol.Chem., 279, 2004
|
|
5A39
 
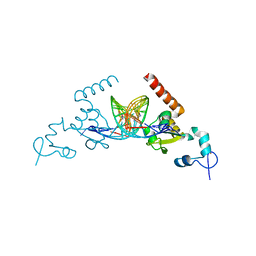 | | Structure of Rad14 in complex with cisplatin containing DNA | | Descriptor: | 5'-D(*DGP*AP*TP*GP*AP*CP*CP*GP*TP*AP*GP*AP*GP)-3', 5'-D(*DTP*GP*AP*TP*GP*AP*CP*CP*GP*TP*AP*GP*AP)-3', Cisplatin, ... | | Authors: | Koch, S.C, Kuper, J, Gasteiger, K.L, Wichlein, N, Strasser, R, Eisen, D, Geiger, S, Schneider, S, Kisker, C, Carell, T. | | Deposit date: | 2015-05-28 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Recognition of Cisplatin and Aaf-Dg Lesion by Rad14 (Xpa).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5LCL
 
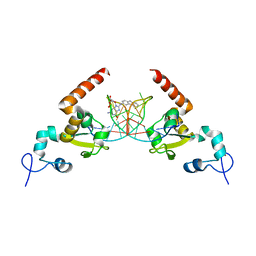 | | STRUCTURE OF the RAD14 DNA-binding domain IN COMPLEX WITH C8-aminofluorene- GUANINE CONTAINING DNA | | Descriptor: | DNA (5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*G)-3'), DNA repair protein RAD14, GCTCTAC(8AF)TCATCA, ... | | Authors: | Schneider, S, Carell, T, Ebert, C, Simon, N. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Recognition of N(2) -Aryl- and C8-Aryl DNA Lesions by the Repair Protein XPA/Rad14.
Chembiochem, 18, 2017
|
|
5LCM
 
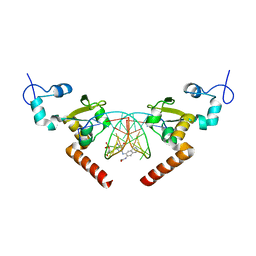 | | STRUCTURE OF the RAD14 DNA-binding domain IN COMPLEX WITH N2-acetylaminonaphtyl- GUANINE CONTAINING DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*CP*TP*AP*CP*(AAN)P*TP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*G)-3'), DNA repair protein RAD14, ... | | Authors: | Schneider, S, Ebert, C, Simon, N, Carell, T. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Recognition of N(2) -Aryl- and C8-Aryl DNA Lesions by the Repair Protein XPA/Rad14.
Chembiochem, 18, 2017
|
|
5A3D
 
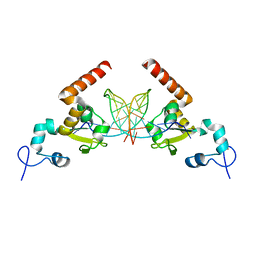 | | Structural insights into the recognition of cisplatin and AAF-dG lesions by Rad14 (XPA) | | Descriptor: | 5'-D(*DG 5IUP*GP*A 5IUP*GP*AP*CP*G 5IUP*AP*GP*AP*DGP*AP)-3', 5'-D(*DTP*CP*TP*CP*TP*AP*C 8FGP*TP*CP*AP*TP*CP*DAP*CP)-3', DNA REPAIR PROTEIN RAD14, ... | | Authors: | Kuper, J, Koch, S.C, Gasteiger, K.L, Wichlein, N, Schneider, S, Kisker, C, Carell, T. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into the Recognition of Cisplatin and Aaf-Dg Lesion by Rad14 (Xpa).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2XRZ
 
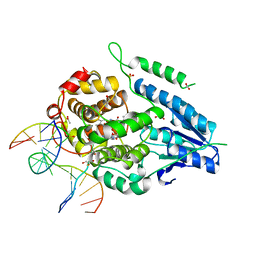 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei in complex with intact CPD-lesion | | Descriptor: | ACETATE ION, COUNTERSTRAND-OLIGONUCLEOTIDE, CPD-COMPRISING OLIGONUCLEOTIDE, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
3C58
 
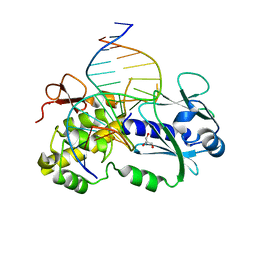 | | Crystal structure of a complex between the wild-type lactococcus lactis Fpg (MutM) and a N7-Benzyl-Fapy-dG containing DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DCP*DTP*DTP*DTP*(SOS)P*DTP*DTP*DTP*DCP*DTP*DCP*DG)-3'), DNA (5'-D(*DGP*DCP*DGP*DAP*DGP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DGP*DA)-3'), DNA glycosylase, ... | | Authors: | Coste, F, Castaing, B, Carell, T. | | Deposit date: | 2008-01-31 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bacterial base excision repair enzyme Fpg recognizes bulky N7-substituted-FapydG lesion via unproductive binding mode
Chem.Biol., 15, 2008
|
|
3CVX
 
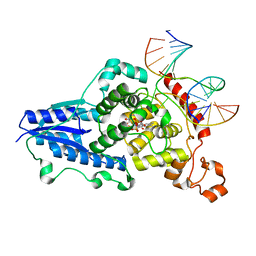 | | Drosophila melanogaster (6-4) photolyase H369M mutant bound to ds DNA with a T-T (6-4) photolesion | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a coenzyme F0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
3CVW
 
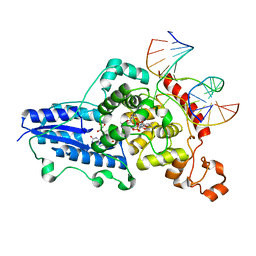 | | Drosophila melanogaster (6-4) photolyase H365N mutant bound to ds DNA with a T-T (6-4) photolesion and cofactor F0 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a cofactor f0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
2XY7
 
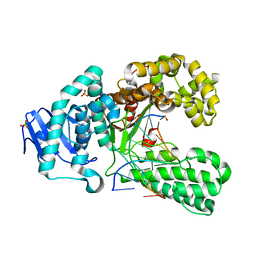 | | Crystal structure of a salicylic aldehyde base in the pre-insertion site of fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | 5'-D(*GP*CP*CP*TP*GP*AP*CP*TP*CP*GP)-3', 5'-D(*SAYP*CP*GP*AP*GP*TP*CP*AP*GP*GP*CP)-3', DNA POLYMERASE I, ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
2Y1I
 
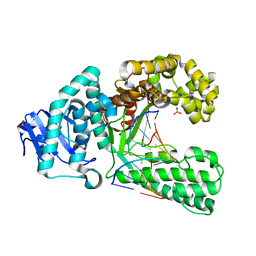 | | Crystal structure of a S-diastereomer analogue of the spore photoproduct in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | 5'-D(*AP*GP*GP*GP*PBTP*THM*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*AP*AP*CP*CP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Heil, K, Schneider, S, Mueller, M, Kneuttinger, A.C, Carell, T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal Structures and Repair Studies Reveal the Identity and the Base-Pairing Properties of the Uv-Induced Spore Photoproduct DNA Lesion.
Chemistry, 17, 2011
|
|
2XRY
 
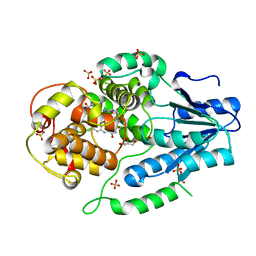 | | X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | DEOXYRIBODIPYRIMIDINE PHOTOLYASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kiontke, S, Geisselbrecht, Y, Pokorny, R, Carell, T, Batschauer, A, Essen, L.O. | | Deposit date: | 2010-09-24 | | Release date: | 2011-09-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of an Archaeal Class II DNA Photolyase and its Complex with Uv-Damaged Duplex DNA.
Embo J., 30, 2011
|
|
2XY5
 
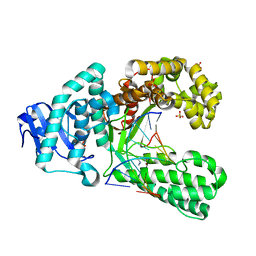 | | Crystal structure of an artificial salen-copper basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 5'-D(*AP*GP*GP*GP*AP*SAYP*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*SAYP*TP*CP*CP*CP*TP)-3', ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
2XY6
 
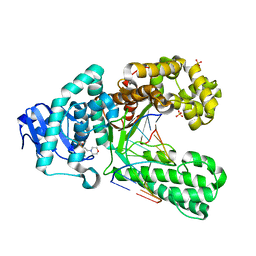 | | Crystal structure of a salicylic aldehyde basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-D(*AP*GP*GP*GP*AP*SAYP*GP*GP*TP*CP)-3', ... | | Authors: | Kaul, C, Mueller, M, Wagner, M, Schneider, S, Carell, T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reversible Bond Formation Enables the Replication and Amplification of a Crosslinking Salen Complex as an Orthogonal Base Pair.
Nature Chem., 3, 2011
|
|
2Y1J
 
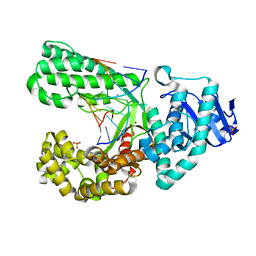 | | CRYSTAL STRUCTURE OF A R-DIASTEREOMER ANALOGUE OF THE SPORE PHOTOPRODUCT IN COMPLEX WITH FRAGMENT DNA POLYMERASE I FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | 5'-D(*AP*GP*GP*GP*QBTP*THM*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*AP*AP*CP*CP*CP*TP)-3', DNA POLYMERASE I, ... | | Authors: | Heil, K, Schneider, S, Mueller, M, Kneuttinger, A.C, Carell, T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-07-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures and Repair Studies Reveal the Identity and the Base-Pairing Properties of the Uv-Induced Spore Photoproduct DNA Lesion.
Chemistry, 17, 2011
|
|
2XGP
 
 | | Yeast DNA polymerase eta in complex with C8-2-acetylaminofluorene containing DNA | | Descriptor: | 5'-D(*CP*8FG*CP*TP*CP*AP*TP*CP*CP*AP*C)-3', 5'-D(*GP*TP*GP*GP*AP*TP*GP*AP*G)-3', CALCIUM ION, ... | | Authors: | Scheider, S, Lammens, K, Schorr, S, Hopfner, K.P, Carell, T. | | Deposit date: | 2010-06-07 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of Replication Blocking and Bypass of Y-Family Polymerase Eta by Bulky Acetylaminofluorene DNA Adducts.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
