6VVZ
 
 | | Mycobacterium tuberculosis RNAP S456L mutant transcription initiation intermediate structure with Sorangicin | | Descriptor: | DNA (62-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVT
 
 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and antibiotic Sorangicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VW0
 
 | | Mycobacterium tuberculosis RNAP S456L mutant open promoter complex | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVS
 
 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with antibiotic Sorangicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Braffman, N, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.112 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVV
 
 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase | | Descriptor: | DNA (26-MER), DNA (31-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVY
 
 | | Mycobacterium tuberculosis WT RNAP transcription open promoter complex with Sorangicin | | Descriptor: | DNA (65-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VVX
 
 | | Mycobacterium tuberculosis WT RNAP transcription initiation intermediate structure with Sorangicin | | Descriptor: | DNA (63-MER), DNA (65-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Lilic, M, Boyaci, H, Chen, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1TH8
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: inhibitory complex with ADP, crystal form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigian, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
1TIL
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA:Poised for phosphorylation complex with ATP, crystal form II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
1THN
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: inhibitory complex with ADP, crystal form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-01 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
1TID
 
 | | Crystal Structures of the ADP and ATP bound forms of the Bacillus Anti-sigma factor SpoIIAB in complex with the Anti-anti-sigma SpoIIAA: Poised for phosphorylation complex with ATP, crystal form I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anti-sigma F factor, Anti-sigma F factor antagonist, ... | | Authors: | Masuda, S, Murakami, K.S, Wang, S, Olson, C.A, Donigan, J, Leon, F, Darst, S.A, Campbell, E.A. | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the ADP and ATP Bound Forms of the Bacillus Anti-sigma Factor SpoIIAB in Complex with the Anti-anti-sigma SpoIIAA.
J.Mol.Biol., 340, 2004
|
|
2FMN
 
 | | Ala177Val mutant of E. coli Methylenetetrahydrofolate Reductase complex with LY309887 | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE, N-[(5-{2-[(6R)-2-AMINO-4-OXO-3,4,5,6,7,8-HEXAHYDROPYRIDO[2,3-D]PYRIMIDIN-6-YL]ETHYL}-2-THIENYL)CARBONYL]-L-GLUTAMIC ACID | | Authors: | Pejchal, R, Campbell, E, Guenther, B.D, Lennon, B.W, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Perturbations in the Ala -> Val Polymorphism of Methylenetetrahydrofolate Reductase: How Binding of Folates May Protect against Inactivation
Biochemistry, 45, 2006
|
|
2FMO
 
 | | Ala177Val mutant of E. coli Methylenetetrahydrofolate Reductase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pejchal, R, Campbell, E, Guenther, B.D, Lennon, B.W, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2006-01-09 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Perturbations in the Ala -> Val Polymorphism of Methylenetetrahydrofolate Reductase: How Binding of Folates May Protect against Inactivation
Biochemistry, 45, 2006
|
|
1HQM
 
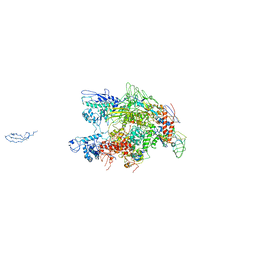 | | CRYSTAL STRUCTURE OF THERMUS AQUATICUS CORE RNA POLYMERASE-INCLUDES COMPLETE STRUCTURE WITH SIDE-CHAINS (EXCEPT FOR DISORDERED REGIONS)-FURTHER REFINED FROM ORIGINAL DEPOSITION-CONTAINS ADDITIONAL SEQUENCE INFORMATION | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Minakhin, L, Bhagat, S, Brunning, A, Campbell, E.A, Darst, S.A, Ebright, R.H, Severinov, K. | | Deposit date: | 2000-12-18 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1OTS
 
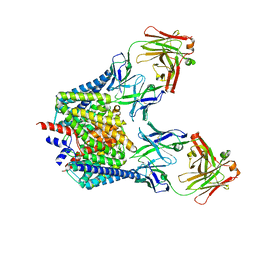 | | Structure of the Escherichia coli ClC Chloride channel and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (heavy chain), Fab fragment (light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-22 | | Release date: | 2003-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1KPK
 
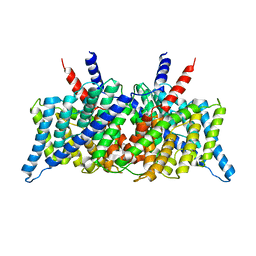 | | Crystal Structure of the ClC Chloride Channel from E. coli | | Descriptor: | putative channel transporter | | Authors: | Dutzler, R, Campbell, E.B, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-12-31 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of
anion selectivity.
Nature, 415, 2002
|
|
1KPL
 
 | | Crystal Structure of the ClC Chloride Channel from S. typhimurium | | Descriptor: | CHLORIDE ION, N-OCTANE, PENTADECANE, ... | | Authors: | Dutzler, R, Campbell, E.B, Cadene, M, Chait, B.T, MacKinnon, R. | | Deposit date: | 2001-12-31 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of a ClC chloride channel at 3.0 A reveals the molecular basis of
anion selectivity.
Nature, 415, 2002
|
|
1OTU
 
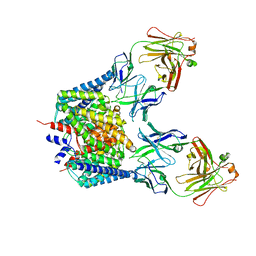 | | Structure of the Escherichia coli ClC Chloride channel E148Q mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
1OTT
 
 | | Structure of the Escherichia coli ClC Chloride channel E148A mutant and Fab Complex | | Descriptor: | CHLORIDE ION, Fab fragment (Heavy chain), Fab fragment (Light chain), ... | | Authors: | Dutzler, R, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2003-03-23 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gating the Selectivity Filter in ClC Chloride Channels
Science, 300, 2003
|
|
