7UW3
 
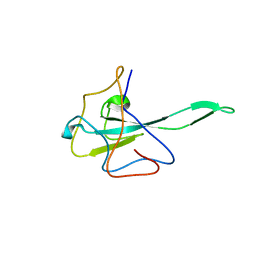 | |
5JK7
 
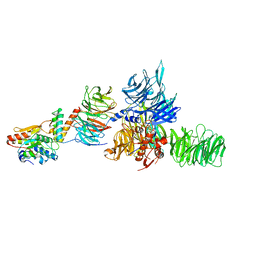 | | The X-ray structure of the DDB1-DCAF1-Vpr-UNG2 complex | | Descriptor: | DNA damage-binding protein 1, Protein VPRBP, Protein Vpr, ... | | Authors: | Calero, G, Ahn, J, Wu, Y. | | Deposit date: | 2016-04-26 | | Release date: | 2016-10-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction.
Nat.Struct.Mol.Biol., 23, 2016
|
|
8FSL
 
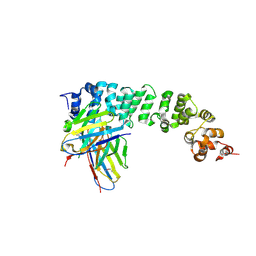 | |
9BVT
 
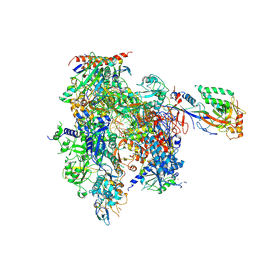 | | RNA Pol II - High Mn(+2) concentration | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Calero, G. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BW0
 
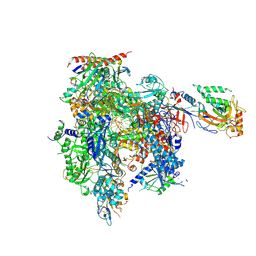 | | RNA Polymerase II - No ATP | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Calero, G. | | Deposit date: | 2024-05-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6XQI
 
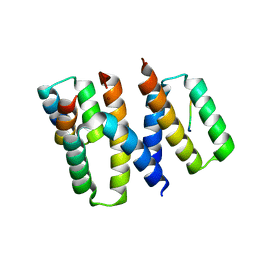 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | Descriptor: | ASN-PRO-LEU-GLU-PHE-LEU, Protein Vpr, UV excision repair protein RAD23 homolog A, ... | | Authors: | Calero, G.C, Wu, Y, Weiss, S.C. | | Deposit date: | 2020-07-09 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
6OI7
 
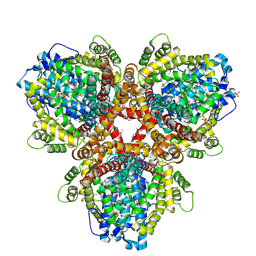 | | Se-Met structure of apo- Escherichia coli dGTPase | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MANGANESE (II) ION, SULFATE ION | | Authors: | Calero, G, Barnes, C.O, Wu, Y. | | Deposit date: | 2019-04-08 | | Release date: | 2019-05-29 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1N52
 
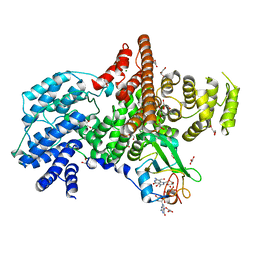 | | Cap Binding Complex | | Descriptor: | 20 kDa nuclear cap binding protein, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, 80 kDa nuclear cap binding protein, ... | | Authors: | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | Deposit date: | 2002-11-04 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
1N54
 
 | | Cap Binding Complex m7GpppG free | | Descriptor: | 20 kDa nuclear cap binding protein, 80 kDa nuclear cap binding protein, GLYCEROL | | Authors: | Calero, G, Wilson, K, Ly, T, Rios-Steiner, J, Clardy, J, Cerione, R. | | Deposit date: | 2002-11-04 | | Release date: | 2003-02-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of m7GpppG binding to the nuclear cap-binding protein complex.
Nat.Struct.Biol., 9, 2002
|
|
1PJA
 
 | | The crystal structure of palmitoyl protein thioesterase-2 reveals the basis for divergent substrate specificities of the two lysosomal thioesterases (PPT1 and PPT2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Palmitoyl-protein thioesterase 2 precursor | | Authors: | Calero, G, Gupta, P, Nonato, M.C, Tandel, S, Biehl, E.R, Hofmann, S.L, Clardy, J. | | Deposit date: | 2003-06-02 | | Release date: | 2003-09-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase-2 (PPT2) reveals the basis for divergent substrate specificities of the two lysosomal thioesterases, PPT1 and PPT2.
J.Biol.Chem., 278, 2003
|
|
7SD4
 
 | | SARS-CoV-2 Nucleocapsid N-terminal domain (N-NTD) protein | | Descriptor: | Nucleoprotein | | Authors: | Sarkar, S, Runge, B, Russell, R.W, Calero, D, Zeinalilathori, S, Quinn, C.M, Lu, M, Calero, G, Gronenborn, A.M, Polenova, T. | | Deposit date: | 2021-09-29 | | Release date: | 2022-06-08 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-Resolution Structure of SARS-CoV-2 Nucleocapsid Protein N-Terminal Domain.
J.Am.Chem.Soc., 144, 2022
|
|
5C3E
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C44
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C4J
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C4X
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Spahr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-18 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
5C4A
 
 | | Crystal structure of a transcribing RNA Polymerase II complex reveals a complete transcription bubble | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Barnes, C.O, Calero, M, Malik, I, Saphr, H, Zhang, Q, Pullara, F, Kaplan, C.D, Calero, G. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble.
Mol.Cell, 59, 2015
|
|
2QRZ
 
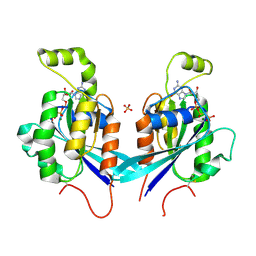 | | Cdc42 bound to GMP-PCP: Induced Fit by Effector is Required | | Descriptor: | Cell division control protein 42 homolog precursor, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Phillips, M.J, Calero, G, Chan, B, Cerione, R.A. | | Deposit date: | 2007-07-30 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effector Proteins Exert an Important Influence on the Signaling-active State of the Small GTPase Cdc42.
J.Biol.Chem., 283, 2008
|
|
8U9X
 
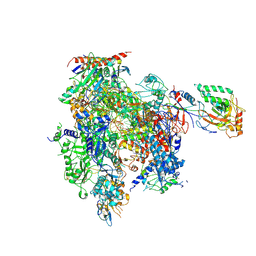 | | STRUCTURAL BASIS OF TRANSCRIPTION: RNA POLYMERASE II SUBSTRATE BINDING AND METAL COORDINATION AT 3.0 A OF T834P MUTANT USING A FREE-ELECTRON LASER | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Arjunan, P, Calero, G, Kaplan, C.D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U9R
 
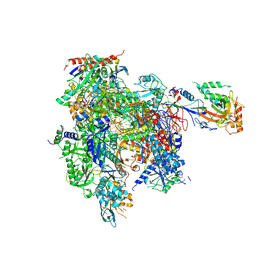 | | STRUCTURAL BASIS OF TRANSCRIPTION: RNA POLYMERASE II SUBSTRATE BINDING AND METAL COORDINATION USING A FREE-ELECTRON LASER | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Arjunan, P, Calero, G, Kaplan, C.D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
3RJ1
 
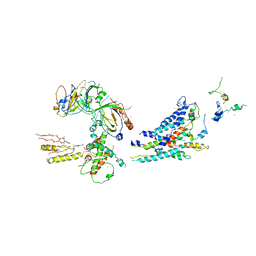 | | Architecture of the Mediator Head module | | Descriptor: | Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, Mediator of RNA polymerase II transcription subunit 18, ... | | Authors: | Imasaki, T, Calero, G, Cai, G, Tsai, K.L, Yamada, K, Cardelli, F, Erdjument-Bromage, H, Tempst, P, Berger, I, Kornberg, G.L, Asturias, F.J, Kornberg, R.D, Takagi, Y. | | Deposit date: | 2011-04-14 | | Release date: | 2011-07-27 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Architecture of the Mediator head module.
Nature, 475, 2011
|
|
4Z8L
 
 | | Crystal structure of DCAF1/SIV-MND VPX/MND SAMHD1 NTD ternary complex | | Descriptor: | Protein VPRBP, SAM domain and HD domain-containing protein, Vpx protein, ... | | Authors: | Koharudin, L.M, Wu, Y, Calero, G, Ahn, J, Gronenborn, A.M. | | Deposit date: | 2015-04-09 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Clade-specific Engagement of SAMHD1 (Sterile alpha Motif and Histidine/Aspartate-containing Protein 1) Restriction Factors by Lentiviral Viral Protein X (Vpx) Virulence Factors.
J.Biol.Chem., 290, 2015
|
|
6OIX
 
 | | Structure of Escherichia coli dGTPase bound to GTP | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OIV
 
 | | XFEL structure of Escherichia coli dGTPase | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MANGANESE (II) ION, SULFATE ION | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6XQJ
 
 | | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A | | Descriptor: | Protein Vpr,UV excision repair protein RAD23 homolog A, ZINC ION | | Authors: | Byeon, I.-J.L, Calero, G, Wu, Y, Byeon, C.H, Gronenborn, A.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of HIV-1 Vpr in complex with the human nucleotide excision repair protein hHR23A.
Nat Commun, 12, 2021
|
|
6OIW
 
 | | Structure of Escherichia coli dGTPase bound to dGTP-1-thiol | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxyguanosinetriphosphate triphosphohydrolase, MAGNESIUM ION, ... | | Authors: | Barnes, C.O, Wu, Y, Calero, G. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | The crystal structure of dGTPase reveals the molecular basis of dGTP selectivity.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
