1MIT
 
 | | RECOMBINANT CUCURBITA MAXIMA TRYPSIN INHIBITOR V (RCMTI-V) (NMR, MINIMIZED AVERAGE STRUCTURE) | | Descriptor: | TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Huang, Y, Liu, J, Prakash, O, Wen, L, Wen, J.J, Huang, J.-K, Krishnamoorthi, R. | | Deposit date: | 1995-10-26 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of recombinant Cucurbita maxima trypsin inhibitor-V determined by NMR spectroscopy.
Biochemistry, 35, 1996
|
|
2LRK
 
 | | Solution Structures of the IIA(Chitobiose)-HPr complex of the N,N'-Diacetylchitobiose | | Descriptor: | N,N'-diacetylchitobiose-specific phosphotransferase enzyme IIA component, Phosphocarrier protein HPr | | Authors: | Cai, M, Jung, Y, Clore, M. | | Deposit date: | 2012-04-06 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the IIAChitobiose-HPr Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia coli Phosphotransferase System.
J.Biol.Chem., 287, 2012
|
|
1WJF
 
 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, 40 STRUCTURES | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
1HYM
 
 | | HYDROLYZED TRYPSIN INHIBITOR (CMTI-V, MINIMIZED AVERAGE NMR STRUCTURE) | | Descriptor: | HYDROLYZED CUCURBITA MAXIMA TRYPSIN INHIBITOR V | | Authors: | Cai, M, Gong, Y, Prakash, O, Krishnamoorthi, R. | | Deposit date: | 1995-06-12 | | Release date: | 1995-09-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Reactive-site hydrolyzed Cucurbita maxima trypsin inhibitor-V: function, thermodynamic stability, and NMR solution structure.
Biochemistry, 34, 1995
|
|
6U6R
 
 | |
6U6P
 
 | |
6U6S
 
 | |
6U6Q
 
 | |
1WJE
 
 | | SOLUTION STRUCTURE OF H12C MUTANT OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE COMPLEXED TO CADMIUM, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CADMIUM ION, HIV-1 INTEGRASE | | Authors: | Cai, M, Gronenborn, A.M, Clore, G.M. | | Deposit date: | 1998-06-11 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the His12 --> Cys mutant of the N-terminal zinc binding domain of HIV-1 integrase complexed to cadmium.
Protein Sci., 7, 1998
|
|
1COR
 
 | |
1CCH
 
 | |
2LOE
 
 | | Structure of the Plasmodium 6-cysteine s48/45 Domain | | Descriptor: | 6-cysteine protein, putative | | Authors: | Cai, M, Arredondo, S.A, Clore, M.G, Miller, L.H, Takayama, Y, Macdonald, N.J, Enderson, E.D, Aravind, L. | | Deposit date: | 2012-01-23 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the Plasmodium 6-cysteine s48/45 domain.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1TIN
 
 | |
6QGN
 
 | | Crystal structure of APT1 bound to 2-Bromopalmitate | | Descriptor: | 2-Bromopalmitic acid, Acyl-protein thioesterase 1 | | Authors: | Marcaida, M.J, Audagnotto, M, Ho, S, Pojer, F, Van der Goot, G, Dal Peraro, M. | | Deposit date: | 2019-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Palmitoylated acyl protein thioesterase APT2 deforms membranes to extract substrate acyl chains.
Nat.Chem.Biol., 2021
|
|
4ZTR
 
 | | Human Aurora A catalytic domain bound to FK1141 | | Descriptor: | 6-({4-[(Z)-{(2Z)-2-[(4-ethylphenyl)imino]-3-methyl-4-oxo-1,3-thiazolidin-5-ylidene}methyl]pyridin-2-yl}amino)pyridine-3-carboxylic acid, Aurora kinase A | | Authors: | Marcaida, M.J, Kilchmann, F, Schick, T, Reymond, J.L. | | Deposit date: | 2015-05-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery of a Selective Aurora A Kinase Inhibitor by Virtual Screening.
J.Med.Chem., 59, 2016
|
|
4ZTQ
 
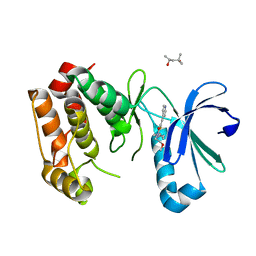 | | Human Aurora A catalytic domain bound to FK932 | | Descriptor: | (2Z,5Z)-2-[(4-ethylphenyl)imino]-3-(2-methoxyethyl)-5-(pyridin-4-ylmethylidene)-1,3-thiazolidin-4-one, (4S)-2-METHYL-2,4-PENTANEDIOL, Aurora kinase A | | Authors: | Marcaida, M.J, Kilchmann, F, Schick, T, Reymond, J.L. | | Deposit date: | 2015-05-14 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Selective Aurora A Kinase Inhibitor by Virtual Screening.
J.Med.Chem., 59, 2016
|
|
2OSX
 
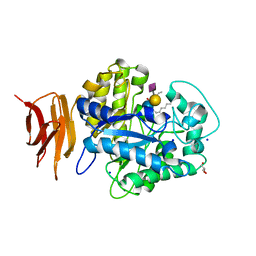 | | Endo-glycoceramidase II from Rhodococcus sp.: Ganglioside GM3 Complex | | Descriptor: | Endoglycoceramidase II, GLYCEROL, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-06 | | Release date: | 2007-02-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural and Mechanistic Analyses of endo-Glycoceramidase II, a Membrane-associated Family 5 Glycosidase in the Apo and GM3 Ganglioside-bound Forms.
J.Biol.Chem., 282, 2007
|
|
2OYL
 
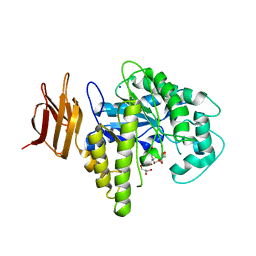 | | Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like imidazole complex | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, Endoglycoceramidase II, GLYCEROL, ... | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structural basis of glycosidase inhibition by five-membered iminocyclitols: the clan a glycoside hydrolase endoglycoceramidase as a model system.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2OYK
 
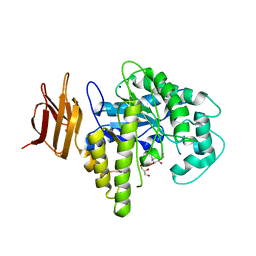 | | Endo-glycoceramidase II from Rhodococcus sp.: cellobiose-like isofagomine complex | | Descriptor: | (3R,4R,5R)-3-hydroxy-5-(hydroxymethyl)piperidin-4-yl beta-D-glucopyranoside, Endoglycoceramidase II, GLYCEROL, ... | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis of glycosidase inhibition by five-membered iminocyclitols: the clan a glycoside hydrolase endoglycoceramidase as a model system.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
4ZTS
 
 | | Human Aurora A catalytic domain bound to FK1142 | | Descriptor: | (2Z,5Z)-2-[(4-ethylphenyl)imino]-3-methyl-5-[(2-{[4-(1H-tetrazol-5-yl)phenyl]amino}pyridin-4-yl)methylidene]-1,3-thiazolidin-4-one, (4S)-2-METHYL-2,4-PENTANEDIOL, Aurora kinase A | | Authors: | Marcaida, M.J, Kilchmann, F, Schick, T, Reymond, J.L. | | Deposit date: | 2015-05-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a Selective Aurora A Kinase Inhibitor by Virtual Screening.
J.Med.Chem., 59, 2016
|
|
3ECQ
 
 | | Endo-alpha-N-acetylgalactosaminidase from Streptococcus pneumoniae: SeMet structure | | Descriptor: | CALCIUM ION, Endo-alpha-N-acetylgalactosaminidase, GLYCEROL, ... | | Authors: | Caines, M.E.C, Zhu, H, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2008-09-01 | | Release date: | 2008-09-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structural basis for T-antigen hydrolysis by Streptococcus pneumoniae: a target for structure-based vaccine design.
J.Biol.Chem., 283, 2008
|
|
2OSW
 
 | | Endo-glycoceramidase II from Rhodococcus sp. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endoglycoceramidase II, SODIUM ION | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-06 | | Release date: | 2007-02-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Mechanistic Analyses of endo-Glycoceramidase II, a Membrane-associated Family 5 Glycosidase in the Apo and GM3 Ganglioside-bound Forms.
J.Biol.Chem., 282, 2007
|
|
2OSY
 
 | |
2OYM
 
 | | Endo-glycoceramidase II from Rhodococcus sp.: five-membered iminocyclitol complex | | Descriptor: | Endoglycoceramidase II, N-{[(2R,3R,4R,5R)-3,4-DIHYDROXY-5-(HYDROXYMETHYL)PYRROLIDIN-2-YL]METHYL}-4-(DIMETHYLAMINO)BENZAMIDE, SODIUM ION | | Authors: | Caines, M.E.C, Strynadka, N.C.J. | | Deposit date: | 2007-02-22 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structural basis of glycosidase inhibition by five-membered iminocyclitols: the clan a glycoside hydrolase endoglycoceramidase as a model system.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
2IHU
 
 | | Carboxyethylarginine synthase from Streptomyces clavuligerus: putative reaction intermediate complex | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-2-[(1Z)-1-HYDROXY-3-(PHOSPHONOOXY)PROP-1-EN-1-YL]-3-{[(4Z)-4-IMINO-2- METHYL-4,5-DIHYDROPYRIMIDIN-5-YL]METHYL}-4-METHYL-1,3-THIAZOL-3-IUM, Carboxyethylarginine synthase, ... | | Authors: | Caines, M.E, Schofield, C.J. | | Deposit date: | 2006-09-27 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and mechanistic studies on N(2)-(2-carboxyethyl)arginine synthase.
Biochem.Biophys.Res.Commun., 385, 2009
|
|
