4W4O
 
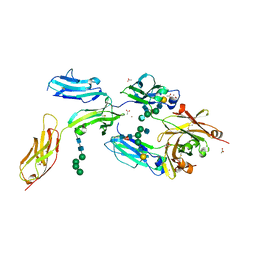 | | High-resolution crystal structure of Fc bound to its human receptor Fc-gamma-RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kiyoshi, M, Tsumoto, K. | | Deposit date: | 2014-08-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Nat Commun, 6, 2015
|
|
6K2G
 
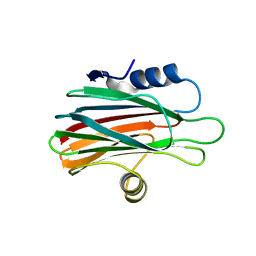 | |
8JOW
 
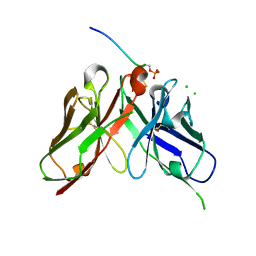 | |
3FZW
 
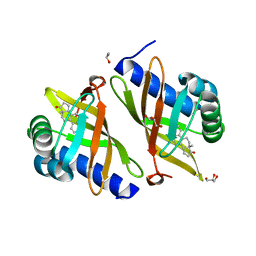 | | Crystal Structure of Ketosteroid Isomerase D40N-D103N from Pseudomonas putida (pKSI) with bound equilenin | | Descriptor: | EQUILENIN, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Caaveiro, J.M.M, Ringe, D, Petsko, G.A. | | Deposit date: | 2009-01-26 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Hydrogen bond coupling in the ketosteroid isomerase active site.
Biochemistry, 48, 2009
|
|
3CPO
 
 | | Crystal structure of ketosteroid isomerase D40N with bound 2-fluorophenol | | Descriptor: | 2-fluorophenol, Delta(5)-3-ketosteroid isomerase | | Authors: | Caaveiro, J.M.M, Pybus, B, Ringe, D, Petsko, G. | | Deposit date: | 2008-03-31 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Testing geometrical discrimination within an enzyme active site: constrained hydrogen bonding in the ketosteroid isomerase oxyanion hole
J.Am.Chem.Soc., 130, 2008
|
|
4W4N
 
 | | Crystal structure of human Fc at 1.80 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kiyoshi, M, Tsumoto, K. | | Deposit date: | 2014-08-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Nat Commun, 6, 2015
|
|
5GWF
 
 | | FraC with GlcNAc(6S) bound | | Descriptor: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, CHLORIDE ION, DELTA-actitoxin-Afr1a, ... | | Authors: | Caaveiro, J.M.M, Tsumoto, K. | | Deposit date: | 2016-09-11 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Haemolytic actinoporins interact with carbohydrates using their lipid-binding module
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 372, 2017
|
|
5JYM
 
 | | Human P-cadherin EC12 with scFv TSP11 bound | | Descriptor: | Antibody scFv TSP11, CALCIUM ION, Cadherin-3 | | Authors: | Caaveiro, J.M.M, Kudo, S, Tsumoto, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Disruption of cell adhesion by an antibody targeting the cell-adhesive intermediate (X-dimer) of human P-cadherin
Sci Rep, 7, 2017
|
|
5JYL
 
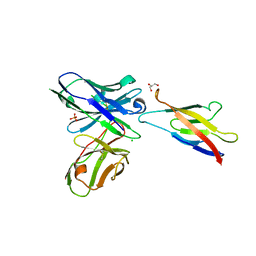 | | Human P-cadherin MEC1 with scFv TSP7 bound | | Descriptor: | CHLORIDE ION, Cadherin-3, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kudo, S, Tsumoto, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Disruption of cell adhesion by an antibody targeting the cell-adhesive intermediate (X-dimer) of human P-cadherin
Sci Rep, 7, 2017
|
|
4WD4
 
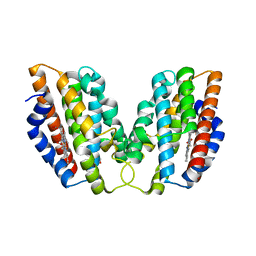 | | Crystal structure of human HO1 H25R | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Caaveiro, J.M.M, Morante, K, Sigala, P, Tsumoto, K. | | Deposit date: | 2014-09-06 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | In-Cell Enzymology To Probe His-Heme Ligation in Heme Oxygenase Catalysis
Biochemistry, 55, 2016
|
|
5X08
 
 | | Crystal structure of broadly neutralizing anti-HIV-1 antibody 4E10, mutant Npro, with peptide bound | | Descriptor: | ACETATE ION, CHLORIDE ION, Envelope glycoprotein gp160, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2017-01-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Functional Contacts between MPER and the Anti-HIV-1 Broadly Neutralizing Antibody 4E10 Extend into the Core of the Membrane
J. Mol. Biol., 429, 2017
|
|
8H63
 
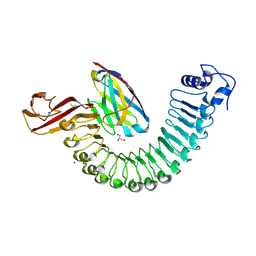 | | Crystal structure of Internalin A from Listeria monocytogenes with nanobody VHH10 bound | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Caaveiro, J.M.M, Nagatoish, S, Tsumoto, K. | | Deposit date: | 2022-10-15 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Anti-InlA single-domain antibodies that inhibit the cell invasion of Listeria monocytogenes.
J.Biol.Chem., 299, 2023
|
|
8H64
 
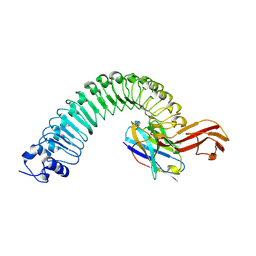 | |
8H62
 
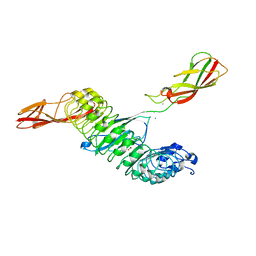 | | Crystal structure of Internalin A from Listeria monocytogenes with human E-cadherin EC12 | | Descriptor: | ACETATE ION, CALCIUM ION, Cadherin-1, ... | | Authors: | Caaveiro, J.M.M, Nagatoish, S, Tsumoto, K. | | Deposit date: | 2022-10-14 | | Release date: | 2023-10-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Anti-InlA single-domain antibodies that inhibit the cell invasion of Listeria monocytogenes.
J.Biol.Chem., 299, 2023
|
|
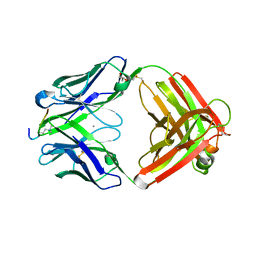
6LDY
 
 | |
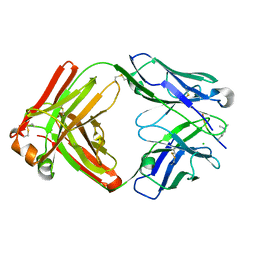
6LDX
 
 | |
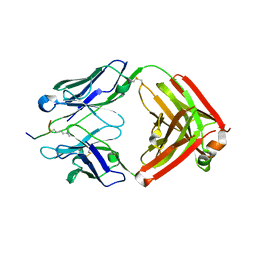
6LDW
 
 | |
6LDV
 
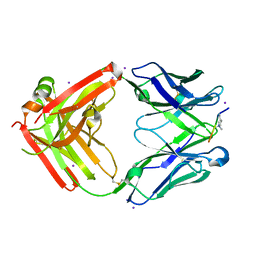 | |
6JB2
 
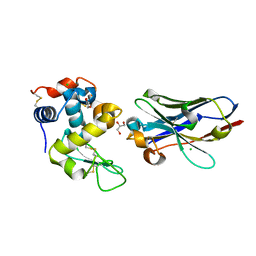 | | Crystal structure of nanobody D3-L11 mutant Y102A in complex with hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
6JB9
 
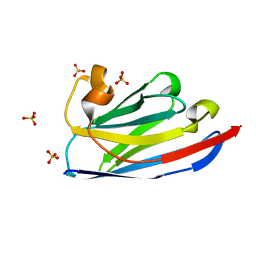 | | Crystal structure of nanobody D3-L11 (unbound form) | | Descriptor: | Nanobody D3-L11, SULFATE ION | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
6JB8
 
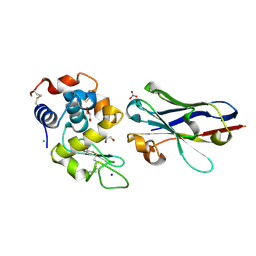 | | Crystal structure of nanobody D3-L11 in complex with hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
6JB5
 
 | | Crystal structure of nanobody D3-L11 mutant Y102A in complex with hen egg-white lysozyme (form II) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Akiba, H, Tsumoto, K. | | Deposit date: | 2019-01-25 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and thermodynamic basis for the recognition of the substrate-binding cleft on hen egg lysozyme by a single-domain antibody.
Sci Rep, 9, 2019
|
|
4WDC
 
 | |
5GHW
 
 | | Crystal structure of broad neutralizing antibody 10E8 with long epitope bound | | Descriptor: | Endogenous retrovirus group K member 8 Env polyprotein, FAB 10E8 HEAVY CHAIN, FAB 10E8 LIGHT CHAIN, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Morante, K, Nieva, J.L, Tsumoto, K. | | Deposit date: | 2016-06-21 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for broad neutralization of HIV-1 through the molecular recognition of 10E8 helical epitope at the membrane interface
Sci Rep, 6, 2016
|
|
7CUE
 
 | | Crystal structure of HID2 bound to human Hemoglobin | | Descriptor: | Amino acid ABC transporter substrate-binding protein, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Caaveiro, J.M.M, Hoshino, M, Tsumoto, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the recognition of human hemoglobin by the Shr protein from Streptococcus pyogenes
To Be Published
|
|
