5T38
 
 | | Crystal Structure of the N-terminal domain of EvdMO1 with SAH bound | | Descriptor: | EvdMO1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCulloch, K.M, Berndt, S, Yamakawa, I, Chen, Q, Loukachevitch, L.V, Starbird, C, Perry, N.A, Iverson, T.M. | | Deposit date: | 2016-08-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1502 Å) | | Cite: | The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Biochemistry, 57, 2018
|
|
5T39
 
 | | Crystal Structure of the N-terminal domain of EvdMO1 in the presence of SAH and D-fucose | | Descriptor: | EvdMO1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCulloch, K.M, Starbird, C.A, Chen, Q, Perry, N.A, Berndt, S, Yamakawa, I, Loukachevitch, L.V, Iverson, T.M. | | Deposit date: | 2016-08-25 | | Release date: | 2017-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1004 Å) | | Cite: | The Structure of the Bifunctional Everninomicin Biosynthetic Enzyme EvdMO1 Suggests Independent Activity of the Fused Methyltransferase-Oxidase Domains.
Biochemistry, 57, 2018
|
|
5YB5
 
 | | The complex crystal structure of VrEH2 mutant M263N with SNO | | Descriptor: | (S)-PARA-NITROSTYRENE OXIDE, Epoxide hydrolase | | Authors: | Li, F.L, Chen, F.F, Chen, Q, Kong, X.D, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-09-03 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation
Acs Catalysis, 8, 2018
|
|
8HW8
 
 | | Crystal structure of Heterodera glycines chitinase 2 D129A/E131A mutant in complex with nodulation factor SmNF-V (C16:2, S) | | Descriptor: | (2~{Z},9~{E})-~{N}-[(2~{R},3~{S},4~{R},5~{S},6~{S})-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]hexadeca-2,9-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, ... | | Authors: | Chen, W, Chen, Q, Wang, D, Yang, Q. | | Deposit date: | 2022-12-29 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structure of Heterodera glycines chitinase 2 D129A/E131A mutant in complex with nodulation factor SmNF-V (C16:2, S)
To Be Published
|
|
8HW7
 
 | | Crystal structure of Heterodera glycines chitinase 2 D129A/E131A mutant in complex with chitopentaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase | | Authors: | Chen, W, Chen, Q, Wang, D, Yang, Q. | | Deposit date: | 2022-12-29 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of Heterodera glycines chitinase 2 D129A/E131A mutant in complex with chitopentaose
To Be Published
|
|
3RQW
 
 | | Crystal structure of acetylcholine bound to a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETYLCHOLINE, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, ... | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
3RQU
 
 | | Crystal structure of a prokaryotic pentameric ligand-gated ion channel, ELIC | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ELIC Pentameric Ligand Gated Ion Channel from Erwinia Chrysanthemi, GLYCEROL | | Authors: | Pan, J.J, Chen, Q, Yoshida, K, Cohen, A, Kong, X.P, Xu, Y, Tang, P. | | Deposit date: | 2011-04-28 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.089 Å) | | Cite: | Structure of the pentameric ligand-gated ion channel ELIC cocrystallized with its competitive antagonist acetylcholine.
Nat Commun, 3, 2012
|
|
5Y6Y
 
 | | The crystal structure of VrEH2 mutant M263N | | Descriptor: | Epoxide hydrolase | | Authors: | Li, F.L, Yu, H.L, Chen, Q, Kong, X.D, Zhou, J.H, Xu, J.H. | | Deposit date: | 2017-08-15 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regioselectivity Engineering of Epoxide Hydrolase: Near-Perfect Enantioconvergence through a Single Site Mutation
Acs Catalysis, 8, 2018
|
|
8HW6
 
 | | Crystal structure of Heterodera glycines chitinase 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Chen, W, Chen, Q, Wang, D, Yang, Q. | | Deposit date: | 2022-12-29 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Crystal structure of Heterodera glycines chitinase 2
To Be Published
|
|
6IEV
 
 | | Crystal structure of a designed protein | | Descriptor: | Designed protein | | Authors: | Han, M, Liao, S, Chen, Q, Liu, H. | | Deposit date: | 2018-09-17 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Selection and analyses of variants of a designed protein suggest importance of hydrophobicity of partially buried sidechains for protein stability at high temperatures.
Protein Sci., 28, 2019
|
|
6J3B
 
 | | Crystal structure of human DHODH in complex with inhibitor 1289 | | Descriptor: | (6R)-1-[3,5-bis(fluoranyl)-4-[2-fluoranyl-5-(hydroxymethyl)phenyl]phenyl]-6-propan-2-yl-6,7-dihydro-5H-benzotriazol-4-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A novel series of human dihydroorotate dehydrogenase inhibitors discovered by in vitro screening: inhibition activity and crystallographic binding mode.
Febs Open Bio, 9, 2019
|
|
6J3C
 
 | | Crystal structure of human DHODH in complex with inhibitor 1291 | | Descriptor: | (6R)-1-[4-[3-(dimethylamino)phenyl]-3,5-bis(fluoranyl)phenyl]-6-propan-2-yl-6,7-dihydro-5H-benzotriazol-4-one, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-01-04 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | A novel series of human dihydroorotate dehydrogenase inhibitors discovered by in vitro screening: inhibition activity and crystallographic binding mode.
Febs Open Bio, 9, 2019
|
|
6JMD
 
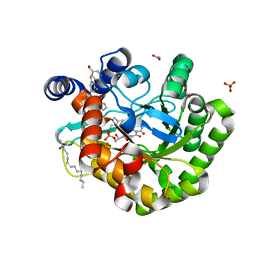 | | Crystal structure of human DHODH in complex with inhibitor 1223 | | Descriptor: | 3-[3,5-bis(fluoranyl)-4-[3-(hydroxymethyl)phenyl]phenyl]benzo[f]benzotriazole-4,9-dione, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-03-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | Bifunctional Naphtho[2,3- d ][1,2,3]triazole-4,9-dione Compounds Exhibit Antitumor Effects In Vitro and In Vivo by Inhibiting Dihydroorotate Dehydrogenase and Inducing Reactive Oxygen Species Production.
J.Med.Chem., 63, 2020
|
|
6JME
 
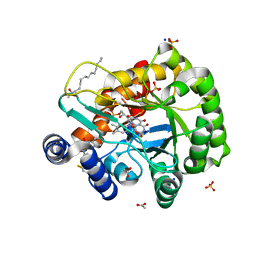 | | Crystal structure of human DHODH in complex with inhibitor 0946 | | Descriptor: | 3-[3,5-bis(fluoranyl)-4-(2-fluorophenyl)phenyl]benzo[f]benzotriazole-4,9-dione, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2019-03-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bifunctional Naphtho[2,3- d ][1,2,3]triazole-4,9-dione Compounds Exhibit Antitumor Effects In Vitro and In Vivo by Inhibiting Dihydroorotate Dehydrogenase and Inducing Reactive Oxygen Species Production.
J.Med.Chem., 63, 2020
|
|
8ZB9
 
 | |
8ZBA
 
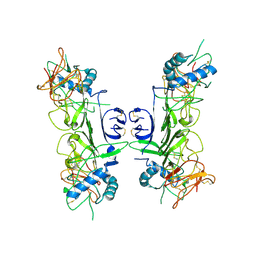 | |
5XHS
 
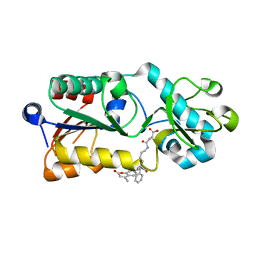 | | Crystal structure of SIRT5 complexed with a fluorogenic small-molecule substrate SuBKA | | Descriptor: | (2S)-2-azanyl-6-[(4-hydroxy-4-oxo-butanoyl)amino]hexanoic acid, 7-AMINO-4-METHYL-CHROMEN-2-ONE, NAD-dependent protein deacylase sirtuin-5, ... | | Authors: | Yu, Y, Li, B, Chen, Q. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions between sirtuins and fluorogenic small-molecule substrates offer insights into inhibitor design
Rsc Adv, 7, 2017
|
|
5ZXH
 
 | | The structure of MT189-tubulin complex | | Descriptor: | 2-(6-fluoro-3-{[(4-methoxyphenyl)methyl]amino}imidazo[1,2-a]pyridin-2-yl)phenol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Li, Z.P, Wang, Y.X, Meng, T, Yang, J.L, Chen, Q. | | Deposit date: | 2018-05-21 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of MT189-Tubulin Complex Provides Insights into Drug Design
Lett.Drug Des.Discovery, 2019
|
|
6KY5
 
 | | Crystal structure of a hydrolase mutant | | Descriptor: | PET hydrolase, SULFATE ION | | Authors: | Cui, Y.L, Chen, Y.C, Liu, X.Y, Dong, S.J, Han, J, Xiang, H, Chen, Q, Liu, H.Y, Han, X, Liu, W.D, Tang, S.Y, Wu, B. | | Deposit date: | 2019-09-16 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Computational redesign of PETase for plasticbiodegradation by GRAPE strategy.
Biorxiv, 2020
|
|
6BCL
 
 | | cryo-EM structure of TRPM4 in apo state with long coiled coil at 3.5 angstrom resolution | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
6BCJ
 
 | | cryo-EM structure of TRPM4 in apo state with short coiled coil at 3.1 angstrom resolution | | Descriptor: | SODIUM ION, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
6BCQ
 
 | | cryo-EM structure of TRPM4 in ATP bound state with long coiled coil at 3.3 angstrom resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
6BCO
 
 | | cryo-EM structure of TRPM4 in ATP bound state with short coiled coil at 2.9 angstrom resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transient receptor potential cation channel subfamily M member 4 | | Authors: | Guo, J, She, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-13 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structures of the calcium-activated, non-selective cation channel TRPM4.
Nature, 552, 2017
|
|
6C9A
 
 | | Cryo-EM structure of mouse TPC1 channel in the PtdIns(3,5)P2-bound state | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,4,6-trihydroxy-3,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | She, J, Guo, J, Chen, Q, Bai, X, Jiang, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-04-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel.
Nature, 556, 2018
|
|
5YWN
 
 | | SsCR_L211H-NADP+ | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Protein induced by osmotic stress | | Authors: | Shang, Y.P, Chen, Q, Yu, H.L, Xu, J.H. | | Deposit date: | 2017-11-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.039 Å) | | Cite: | Attenuated substrate inhibition of a haloketone reductase via structure-guided loop engineering.
J.Biotechnol., 308, 2020
|
|
