3QD0
 
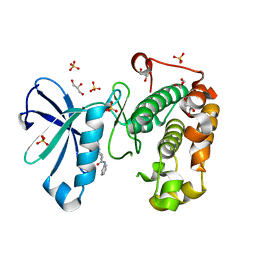 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with (2R,5S)-1-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-N-phenyl-3-piperidinecarboxamide | | Descriptor: | (3S,6R)-1-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-6-methyl-N-phenylpiperidine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
2AYR
 
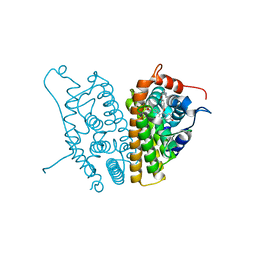 | | A SERM Designed for the Treatment of Uterine Leiomyoma with Unique Tissue Specificity for Uterus and Ovaries in Rats | | Descriptor: | 6-(4-METHYLSULFONYL-PHENYL)-5-[4-(2-PIPERIDIN-1-YLETHOXY)PHENOXY]NAPHTHALEN-2-OL, Estrogen receptor | | Authors: | Hummel, C.W, Geiser, A.G, Bryant, H.U, Cohen, I.R, Dally, R.D, Fong, K.C, Frank, S.A, Hinklin, R, Jones, S.A, Lewis, G, McCann, D.J, Shepherd, T.A, Tian, H, Rudman, D.G, Wallace, O.B, Wang, Y, Dodge, J.A. | | Deposit date: | 2005-09-07 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A selective estrogen receptor modulator designed for the treatment of uterine leiomyoma with unique tissue specificity for uterus and ovaries in rats
J.Med.Chem., 48, 2005
|
|
5WQ0
 
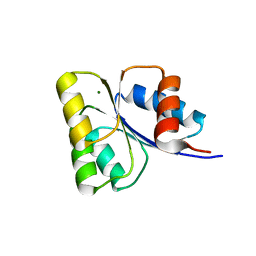 | | Receiver domain of Spo0A from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Stage 0 sporulation protein | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-11-22 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Crystal structure of the inactive state of the receiver domain of Spo0A from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
J. Microbiol., 55, 2017
|
|
2BAY
 
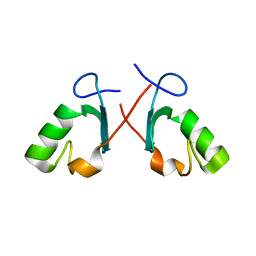 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
1H6F
 
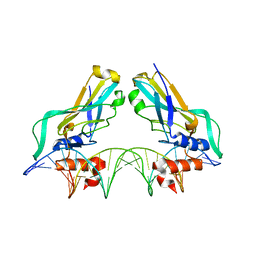 | | Human TBX3, a transcription factor responsible for ulnar-mammary syndrome, bound to a palindromic DNA site | | Descriptor: | 5'-D(*TP*AP*AP*TP*TP*TP*CP*AP*CP*AP*CP*CP*TP* AP*GP*GP*TP*GP*TP*GP*AP*AP*AP*T)-3', MAGNESIUM ION, T-BOX TRANSCRIPTION FACTOR TBX3 | | Authors: | Coll, M, Muller, C.W. | | Deposit date: | 2001-06-13 | | Release date: | 2002-04-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the DNA-Bound T-Box Domain of Human Tbx3, a Transcription Factor Responsible for Ulnar- Mammary Syndrome
Structure, 10, 2002
|
|
1D8A
 
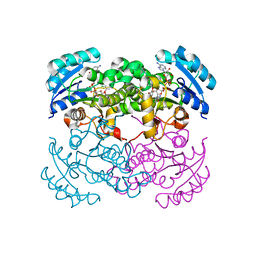 | | E. COLI ENOYL REDUCTASE/NAD+/TRICLOSAN COMPLEX | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Levy, C.W, Roujeinikova, A, Sedelnikova, S, Baker, P.J, Stuitje, A.R, Slabas, A.R, Rice, D.W, Rafferty, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 1999-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of triclosan activity.
Nature, 398, 1999
|
|
1CMY
 
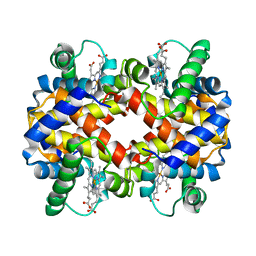 | | THE MUTATION BETA99 ASP-TYR STABILIZES Y-A NEW, COMPOSITE QUATERNARY STATE OF HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN YPSILANTI (CARBONMONOXY) (ALPHA CHAIN), HEMOGLOBIN YPSILANTI (CARBONMONOXY) (BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Smith, F.R, Lattman, E.E, Carter Junior, C.W. | | Deposit date: | 1992-09-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The mutation beta 99 Asp-Tyr stabilizes Y--a new, composite quaternary state of human hemoglobin.
Proteins, 10, 1991
|
|
1DKM
 
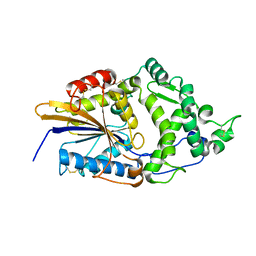 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-02 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKQ
 
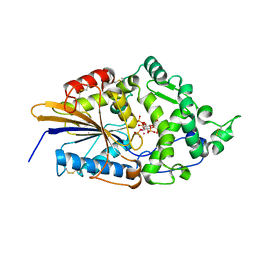 | | CRYSTAL STRUCTURE OF PHYTATE COMPLEX ESCHERICHIA COLI PHYTASE AT PH 5.0. PHYTATE IS BOUND WITH ITS 3-PHOSPHATE IN THE ACTIVE SITE. HG2+ CATION ACTS AS AN INTERMOLECULAR BRIDGE | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, MERCURY (II) ION, PHYTASE | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DKO
 
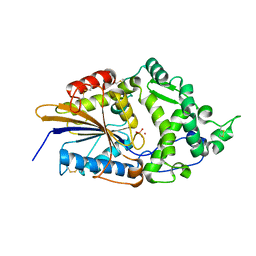 | | CRYSTAL STRUCTURE OF TUNGSTATE COMPLEX OF ESCHERICHIA COLI PHYTASE AT PH 6.6 WITH TUNGSTATE BOUND AT THE ACTIVE SITE AND WITH HG2+ CATION ACTING AS AN INTERMOLECULAR BRIDGE | | Descriptor: | MERCURY (II) ION, PHYTASE, TUNGSTATE(VI)ION | | Authors: | Lim, D, Golovan, S, Forsberg, C.W, Jia, Z. | | Deposit date: | 1999-12-08 | | Release date: | 2000-08-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structures of Escherichia coli phytase and its complex with phytate.
Nat.Struct.Biol., 7, 2000
|
|
1DG4
 
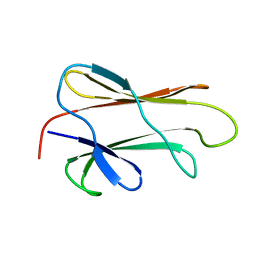 | | NMR STRUCTURE OF THE SUBSTRATE BINDING DOMAIN OF DNAK IN THE APO FORM | | Descriptor: | DNAK | | Authors: | Pellecchia, M, Montgomery, D.L, Stevens, S.Y, Van der Kooi, C.W, Feng, H, Gierasch, L.M, Zuiderweg, E.R.P. | | Deposit date: | 1999-11-23 | | Release date: | 1999-12-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural insights into substrate binding by the molecular chaperone DnaK.
Nat.Struct.Biol., 7, 2000
|
|
1DKL
 
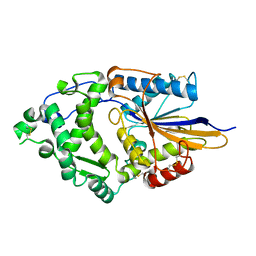 | |
6LVO
 
 | |
6M0Y
 
 | | KR-12 analog derived from human LL-37 | | Descriptor: | LYS-ARG-ILE-VAL-LYS-ARG-ILE-LYS-LYS-TRP-LEU-ARG | | Authors: | Yun, H, Min, H.J, Lee, C.W. | | Deposit date: | 2020-02-24 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Bactericidal Activity of KR-12 Analog Derived from Human LL-37 as a Potential Cosmetic Preservative
To Be Published
|
|
6N55
 
 | | Crystal structure of human uridine-cytidine kinase 2 complexed with 2'-azidouridine | | Descriptor: | 2'-azido-2'-deoxyuridine, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Cuthbert, B.J, Nainar, S, Spitale, R.C, Goulding, C.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.085 Å) | | Cite: | An optimized chemical-genetic method for cell-specific metabolic labeling of RNA.
Nat.Methods, 17, 2020
|
|
6N54
 
 | | Crystal structure of human uridine-cytidine kinase 2 complexed with 2'-azidocytidine monophosphate | | Descriptor: | 2'-azido-2'-deoxycytidine 5'-(dihydrogen phosphate), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuthbert, B.J, Nainar, S, Spitale, R.C, Goulding, C.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | An optimized chemical-genetic method for cell-specific metabolic labeling of RNA.
Nat.Methods, 17, 2020
|
|
6N53
 
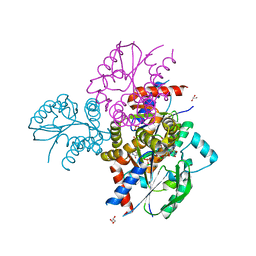 | | Crystal structure of human uridine-cytidine kinase 2 complexed with 2'-azidouridine monophosphate | | Descriptor: | 2'-deoxy-2'-triaza-1,2-dien-2-ium-1-yl-uridine-5'-monophosphate, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuthbert, B.J, Nainar, S, Spitale, R.C, Goulding, C.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An optimized chemical-genetic method for cell-specific metabolic labeling of RNA.
Nat.Methods, 17, 2020
|
|
6OU9
 
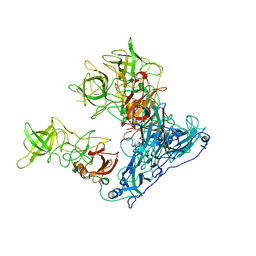 | | Asymmetric focused reconstruction of human norovirus GI.7 Houston strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Major capsid protein | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-04 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OUT
 
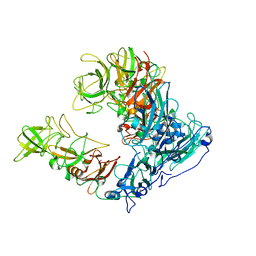 | | Asymmetric focused reconstruction of human norovirus GI.1 Norwalk strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Capsid protein VP1 | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OI4
 
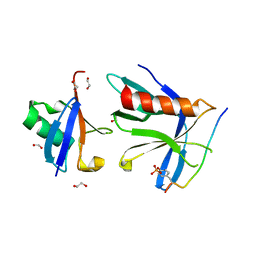 | | RPN13 (19-132)-RPN2 (940-952) pY950-Ub complex | | Descriptor: | 1,2-ETHANEDIOL, 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, ... | | Authors: | Hemmis, C.W, Hill, C.P. | | Deposit date: | 2019-04-08 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Phosphorylation of Tyr-950 in the proteasome scaffolding protein RPN2 modulates its interaction with the ubiquitin receptor RPN13.
J.Biol.Chem., 294, 2019
|
|
5Y9I
 
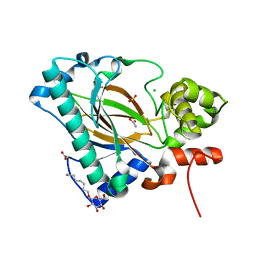 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with Co(II) | | Descriptor: | ACETATE ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H. | | Deposit date: | 2017-08-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Biochemical and structural insights of Fe(II)/alpha-ketoglutarate/O2-dependent dioxygenase, KdoO from Methylacidiphilum infernorum V4
To Be Published
|
|
5Y9X
 
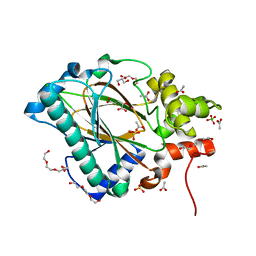 | | Crystal structure of the Kdo hydroxylase KdoO, a non-heme Fe(II) alphaketoglutarate dependent dioxygenase in complex with alphaketoglutarate and coblat(II) | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CHLORIDE ION, ... | | Authors: | Chung, H.S, Pemble, C.W, Joo, S.H, Raetz, C.R. | | Deposit date: | 2017-08-29 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Biochemical and structural insights of Fe(II)/alpha-ketoglutarate/O2-dependent dioxygenase, KdoO from Methylacidiphilum infernorum V4
To Be Published
|
|
2R5A
 
 | |
2R5M
 
 | |
2R57
 
 | |
