1U99
 
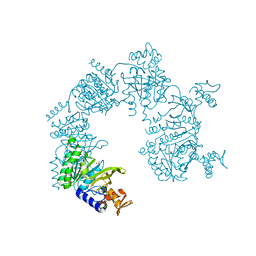 | |
6F76
 
 | | Antibody derived (Abd-8) small molecule binding to KRAS. | | 分子名称: | 4-(2,3-dihydro-1,4-benzodioxin-5-yl)-~{N}-[3-[(dimethylamino)methyl]phenyl]-2-methoxy-aniline, GTPase KRas, MAGNESIUM ION, ... | | 著者 | Bery, N, Cruz-Migoni, A, Quevedo, C.E, Phillips, S.V.E, Carr, S, Rabbitts, T.H. | | 登録日 | 2017-12-07 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions.
Elife, 7, 2018
|
|
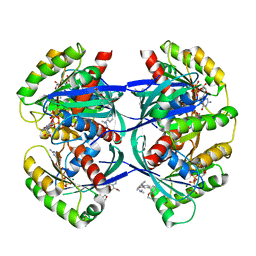
6F94
 
 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(3-methyphenyl)amino]-N-(3-methyphenyl)pyridine-3-carboxamide | | 分子名称: | 6-(ethylcarbamoylamino)-~{N}-(3-methylphenyl)-4-[(3-methylphenyl)amino]pyridine-3-carboxamide, DNA gyrase subunit B | | 著者 | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | 登録日 | 2017-12-14 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
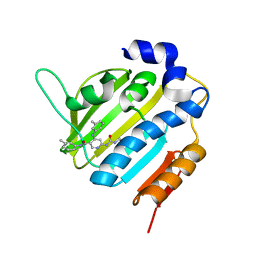
6F9H
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-S-alpha-D-glucopyranosyl-(1,4-dideoxy-4-thio-nojirimycin) | | 分子名称: | 1,4-dideoxy-4-thio-nojirimycin, Beta-amylase, CHLORIDE ION, ... | | 著者 | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | 登録日 | 2017-12-14 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
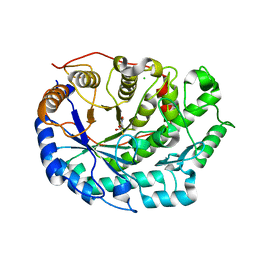
6F9L
 
 | | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose | | 分子名称: | Beta-amylase, CHLORIDE ION, alpha-D-glucopyranose-(1-4)-3-deoxy-3-fluoro-alpha-D-glucopyranose | | 著者 | Tantanarat, K, Stevenson, C.E.M, Rejzek, M, Lawson, D.M, Field, R.A. | | 登録日 | 2017-12-14 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose
To be published
|
|
6F39
 
 | | C1r homodimer CUB1-EGF-CUB2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1r subcomponent, ... | | 著者 | Almitairi, J.O.M, Venkatraman Girija, U, Furze, C.M, Simpson-Gray, X, Badakshi, F, Marshall, J.E, Mitchell, D.A, Moody, P.C.E, Wallis, R. | | 登録日 | 2017-11-28 | | 公開日 | 2018-01-24 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (5.801 Å) | | 主引用文献 | Structure of the C1r-C1s interaction of the C1 complex of complement activation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1Q09
 
 | | Crystal structure of the Zn(II) form of E. coli ZntR, a zinc-sensing transcriptional regulator (space group I4122) | | 分子名称: | SULFATE ION, ZINC ION, Zn(II)-responsive regulator of zntA | | 著者 | Changela, A, Chen, K, Xue, Y, Holschen, J, Outten, C.E, O'Halloran, T.V, Mondragon, A. | | 登録日 | 2003-07-15 | | 公開日 | 2003-09-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR
Science, 301, 2003
|
|
1Q5Z
 
 | | Crystal Structure of the C-terminal Actin Binding Domain of Salmonella Invasion Protein A (SipA) | | 分子名称: | SipA | | 著者 | Stebbins, C.E, Lilic, M, Galkin, V.E, Orlova, A, VanLoock, M.S, Egelman, E.H. | | 登録日 | 2003-08-11 | | 公開日 | 2003-10-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Salmonella SipA polymerizes actin by stapling filaments with nonglobular protein arms.
Science, 301, 2003
|
|
1S1T
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with UC-781 | | 分子名称: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, PHOSPHATE ION, Reverse transcriptase | | 著者 | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | 登録日 | 2004-01-07 | | 公開日 | 2004-06-29 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
6F9J
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-O-alpha-D-mannopyranosyl-(1-deoxynojirimycin) | | 分子名称: | 1-DEOXYNOJIRIMYCIN, Beta-amylase, CHLORIDE ION, ... | | 著者 | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | 登録日 | 2017-12-14 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
7U5J
 
 | |
7U5M
 
 | | Cryo-EM Structure of GAPDH | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Morgan, C.E, Zhang, Z, Yu, E.W. | | 登録日 | 2022-03-02 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.28 Å) | | 主引用文献 | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
7U5H
 
 | |
7U5K
 
 | | Cryo-EM Structure of DPYSL2 | | 分子名称: | Dihydropyrimidinase-related protein 2 | | 著者 | Morgan, C.E, Yu, E.W. | | 登録日 | 2022-03-02 | | 公開日 | 2022-12-14 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (2.78 Å) | | 主引用文献 | Toward structural-omics of the bovine retinal pigment epithelium.
Cell Rep, 41, 2022
|
|
7U5N
 
 | |
7U5I
 
 | |
7U5L
 
 | |
6FA1
 
 | | Antibody derived (Abd-4) small molecule binding to KRAS. | | 分子名称: | 2-[4-[[(3~{R})-2,3-dihydro-1,4-benzodioxin-3-yl]methylcarbamoyl]phenoxy]ethyl-dimethyl-azanium, GTPase KRas, MAGNESIUM ION, ... | | 著者 | Quevedo, C.E, Cruz-Migoni, A, Ehebauer, M.T, Carr, S.B, Phillips, S.V.E, Rabbitts, T.H. | | 登録日 | 2017-12-15 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
1BTQ
 
 | | THE SOLUTION STRUCTURES OF THE FIRST AND SECOND TRANSMEMBRANE-SPANNING SEGMENTS OF BAND 3 | | 分子名称: | BAND 3 ANION TRANSPORT PROTEIN | | 著者 | Gargaro, A.R, Bloomberg, G.B, Dempsey, C.E, Murray, M, Tanner, M.J.A. | | 登録日 | 1994-08-03 | | 公開日 | 1994-11-30 | | 最終更新日 | 2022-02-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structures of the first and second transmembrane-spanning segments of band 3.
Eur.J.Biochem., 221, 1994
|
|
1NDU
 
 | |
1S73
 
 | | Crystal Structure of Mesopone Cytochrome c Peroxidase (R-isomer) [MpCcP-R] | | 分子名称: | Cytochrome c peroxidase, mitochondrial, FE-(4-MESOPORPHYRINONE)-R-ISOMER | | 著者 | Bhaskar, B, Immoos, C.E, Sulc, F, Choen, M.S, Farmer, P.J, Poulos, T.L. | | 登録日 | 2004-01-28 | | 公開日 | 2005-06-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Crystal structures of reduced, resting and NO-bound states of mesopone cytochorme c peroxidase (MpCcP) (R-isomer)
To be Published
|
|
1C7T
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT E540D COMPLEXED WITH DI-N ACETYL-D-GLUCOSAMINE (CHITOBIASE) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | 著者 | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | 登録日 | 2000-03-17 | | 公開日 | 2000-09-20 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
1S1V
 
 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with TNK-651 | | 分子名称: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, Reverse transcriptase | | 著者 | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | 登録日 | 2004-01-07 | | 公開日 | 2004-06-29 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1SBM
 
 | | Crystal Structure of Reduced Mesopone cytochrome c peroxidase (R-isomer) | | 分子名称: | Cytochrome c peroxidase, mitochondrial, FE-(4-MESOPORPHYRINONE)-R-ISOMER | | 著者 | Bhaskar, B, Immoos, C.E, Sulc, F, Cohen, M.S, Farmer, P.J, Poulos, T.L. | | 登録日 | 2004-02-10 | | 公開日 | 2005-06-14 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Crystal Structures of Resting (Fe3+), Reduced (Fe2+) and Reduced-NO adduct of Mesopone cytochrome c peroxidase (MpCcP) - R-isomer
To be Published
|
|
1C7S
 
 | | BETA-N-ACETYLHEXOSAMINIDASE MUTANT D539A COMPLEXED WITH DI-N-ACETYL-BETA-D-GLUCOSAMINE (CHITOBIASE) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE, SULFATE ION | | 著者 | Prag, G, Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K, Oppenheim, A.B. | | 登録日 | 2000-03-14 | | 公開日 | 2000-09-20 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of chitobiase mutants complexed with the substrate Di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540.
J.Mol.Biol., 300, 2000
|
|
