7Z1J
 
 | | Escherichia coli periplasmic phytase AppA, complex with phosphate | | Descriptor: | Acidphosphatase, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2Y
 
 | | Escherichia coli periplasmic phytase AppA T305E mutant, complex with myo-inositol hexakissulfate | | Descriptor: | Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, NICKEL (II) ION | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2S
 
 | | Escherichia coli periplasmic phytase AppA, complex with myo-inositol hexakissulfate | | Descriptor: | Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, NICKEL (II) ION, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z32
 
 | | Escherichia coli periplasmic phytase AppA D304A mutant, phosphohistidine intermediate | | Descriptor: | Acidphosphatase, NICKEL (II) ION | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2T
 
 | | Escherichia coli periplasmic phytase AppA D304A mutant, complex with myo-inositol hexakissulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
8ERA
 
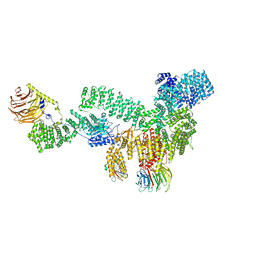 | | RMC-5552 in complex with mTORC1 and FKBP12 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-5,9,27-trihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, 1-[6-{[(3M)-4-amino-3-(2-amino-1,3-benzoxazol-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]methyl}-3,4-dihydroisoquinolin-2(1H)-yl]-3-hydroxypropan-1-one, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
8ER6
 
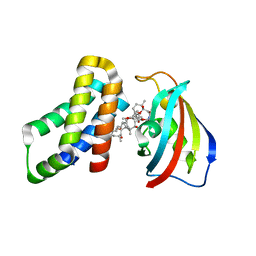 | | FKBP12-FRB in Complex with Compound 11 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-5,9,27-trihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-10,21-dimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, 1,2-ETHANEDIOL, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
8ER7
 
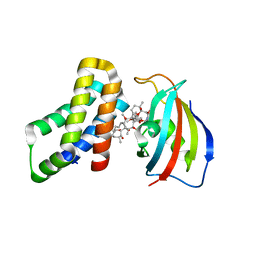 | | FKBP12-FRB in Complex with Compound 12 | | Descriptor: | (3S,5R,6R,7E,9R,10R,12R,14S,15E,17E,19E,21S,23S,26R,27R,30R,34aS)-9,27-dihydroxy-3-{(2R)-1-[(1S,3R,4R)-4-hydroxy-3-methoxycyclohexyl]propan-2-yl}-5,10,21-trimethoxy-6,8,12,14,20,26-hexamethyl-5,6,9,10,12,13,14,21,22,23,24,25,26,27,32,33,34,34a-octadecahydro-3H-23,27-epoxypyrido[2,1-c][1,4]oxazacyclohentriacontine-1,11,28,29(4H,31H)-tetrone, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Tomlinson, A.C.A, Yano, J.K. | | Deposit date: | 2022-10-11 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1-Activated Tumors.
J.Med.Chem., 66, 2023
|
|
7ZBG
 
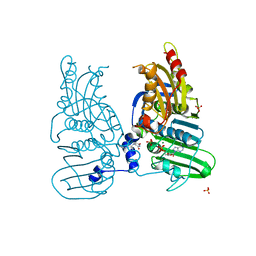 | | Human Topoisomerase II Beta ATPase ADP | | Descriptor: | 4-[(2S,3R)-3-[3,5-bis(oxidanylidene)piperazin-1-ium-1-yl]butan-2-yl]piperazin-4-ium-2,6-dione, ADENOSINE-5'-DIPHOSPHATE, DNA topoisomerase 2-beta, ... | | Authors: | Ling, E.M, Basle, A, Cowell, I.G, Blower, T.R, Austin, C.A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A comprehensive structural analysis of the ATPase domain of human DNA topoisomerase II beta bound to AMPPNP, ADP, and the bisdioxopiperazine, ICRF193.
Structure, 30, 2022
|
|
7Z3V
 
 | | Escherichia coli periplasmic phytase AppA D304E mutant, complex with myo-inositol hexakissulfate | | Descriptor: | Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, POTASSIUM ION | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
6FIK
 
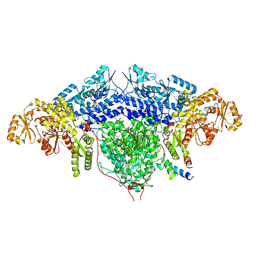 | | ACP2 crosslinked to the KS of the loading/condensing region of the CTB1 PKS | | Descriptor: | Polyketide synthase | | Authors: | Herbst, D.A, Huitt-Roehl, C.R, Jakob, R.P, Townsend, C.A, Maier, T. | | Deposit date: | 2018-01-18 | | Release date: | 2018-03-21 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | The structural organization of substrate loading in iterative polyketide synthases.
Nat. Chem. Biol., 14, 2018
|
|
7Z2A
 
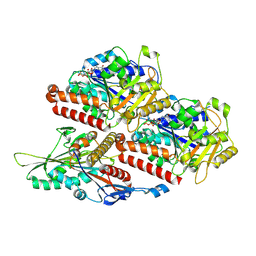 | | P. berghei kinesin-8B motor domain in no nucleotide state bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-8, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
8EW8
 
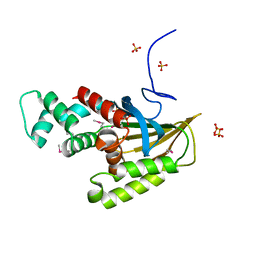 | | Crystal structure of Saccharomyces cerevisiae Altered Inheritance rate of Mitochondria protein 18 (AIM18p) R123A mutant | | Descriptor: | Altered inheritance of mitochondria protein 18, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Schmitz, J.M, Smith, R.W, Pagliarini, D.J. | | Deposit date: | 2022-10-21 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Aim18p and Aim46p are chalcone isomerase domain-containing mitochondrial hemoproteins in Saccharomyces cerevisiae.
J.Biol.Chem., 299, 2023
|
|
7Z2B
 
 | | P. berghei kinesin-8B motor domain in AMPPNP state bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-8, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
8EW9
 
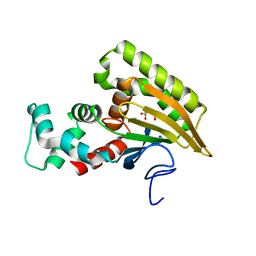 | | Crystal structure of Saccharomyces cerevisiae Altered Inheritance rate of Mitochondria protein 46 (AIM46p) | | Descriptor: | 2-OXOGLUTARIC ACID, Altered inheritance of mitochondria protein 46, mitochondrial | | Authors: | Bingman, C.A, Schmitz, J.M, Smith, R.W, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2022-10-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aim18p and Aim46p are chalcone isomerase domain-containing mitochondrial hemoproteins in Saccharomyces cerevisiae.
J.Biol.Chem., 299, 2023
|
|
7N1H
 
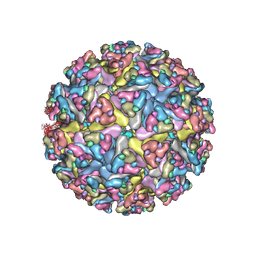 | | CryoEM structure of Venezuelan equine encephalitis virus VLP in complex with the LDLRAD3 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Capsid, ... | | Authors: | Basore, K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor.
Nature, 598, 2021
|
|
6G09
 
 | | Crystal Structure of a GH8 xylobiose complex from Teredinibacter turnerae | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 8 domain protein, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7N1I
 
 | | CryoEM structure of Venezuelan equine encephalitis virus VLP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid, E1 envelope glycoprotein, ... | | Authors: | Basore, K, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-27 | | Release date: | 2021-10-13 | | Last modified: | 2021-11-10 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor.
Nature, 598, 2021
|
|
7NBA
 
 | | Plasmodium falciparum kinesin-5 motor domain bound to AMPPNP, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin motor domain-containing protein,Kinesin motor domain-containing protein, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
8EIZ
 
 | | Cryo-EM structure of squid sensory receptor CRB1 | | Descriptor: | N-benzyl-2-(2,6-dimethylanilino)-N,N-diethyl-2-oxoethan-1-aminium, Squid sensory receptor CRB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, van Giesen, L, Kilian, P.B, Bai, X, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
7NB8
 
 | | Plasmodium falciparum kinesin-5 motor domain without nucleotide, complexed with 14 protofilament microtubule. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Kinesin-5, MAGNESIUM ION, ... | | Authors: | Cook, A.D, Roberts, A, Atherton, J, Tewari, R, Topf, M, Moores, C.A. | | Deposit date: | 2021-01-25 | | Release date: | 2021-10-13 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of a microtubule-bound parasite kinesin motor and implications for its mechanism and inhibition.
J.Biol.Chem., 297, 2021
|
|
8EIS
 
 | | Cryo-EM structure of octopus sensory receptor CRT1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Octopus sensory receptor | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
5N6W
 
 | | Retinoschisin R141H Mutant | | Descriptor: | Retinoschisin | | Authors: | Ramsay, E.P, Collins, R.F, Owens, T.W, Siebert, C.A, Jones, R.P.O, Roseman, A, Wang, T, Baldock, C. | | Deposit date: | 2017-02-16 | | Release date: | 2017-04-12 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of X-linked retinoschisis mutations reveals distinct classes which differentially effect retinoschisin function
Human Molecular Genetics, 25, 2016
|
|
8SSF
 
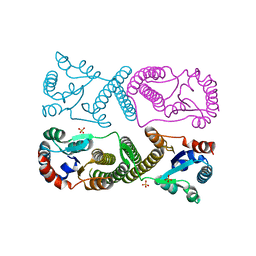 | | Minimal protein-only/RNA-free Ribonuclease P from Hydrogenobacter thermophilus | | Descriptor: | RNA-free ribonuclease P, SULFATE ION | | Authors: | Mendoza, J, Mallik, L, Wilhelm, C.A, Koutmos, M. | | Deposit date: | 2023-05-08 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial RNA-free RNase P: Structural and functional characterization of multiple oligomeric forms of a minimal protein-only ribonuclease P.
J.Biol.Chem., 299, 2023
|
|
8SSG
 
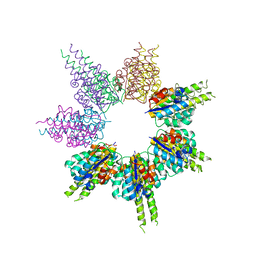 | |
