3ZK8
 
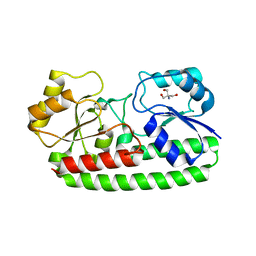 | | CRYSTAL STRUCTURE OF PNEUMOCOCCAL SURFACE ANTIGEN PSAA E205Q IN THE METAL-FREE, OPEN STATE | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | Authors: | Counago, R.M, Ween, M.P, Bajaj, M, Zuegg, J, Cooper, M.A, McEwan, A.G, Paton, J.C, Kobe, B, McDevitt, C.A. | | Deposit date: | 2013-01-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imperfect coordination chemistry facilitates metal ion release in the Psa permease.
Nat. Chem. Biol., 10, 2014
|
|
2KFN
 
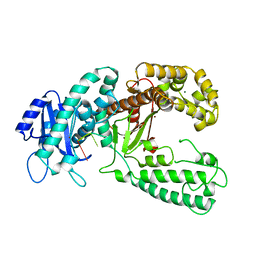 | | KLENOW FRAGMENT WITH BRIDGING-SULFUR SUBSTRATE AND MANGANESE | | Descriptor: | 5'-D(*GP*CP*TP*TP*AP*(US1)P*G)-3', KLENOW FRAGMENT OF DNA POLYMERASE I, MAGNESIUM ION, ... | | Authors: | Brautigam, C.A, Sun, S, Piccirilli, J.A, Steitz, T.A. | | Deposit date: | 1998-07-01 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Biochemistry, 38, 1999
|
|
5J8D
 
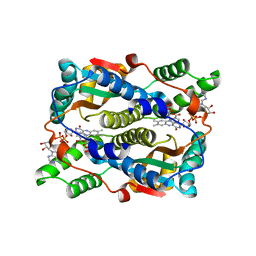 | | Structure of nitroreductase from E. cloacae complexed with nicotinic acid adenine dinucleotide | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINIC ACID ADENINE DINUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Haynes, C.A, Koder, R.L, Miller, A.F, Rodgers, D.W. | | Deposit date: | 2016-04-07 | | Release date: | 2017-05-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism-Informed Refinement Reveals Altered Substrate-Binding Mode for Catalytically Competent Nitroreductase.
Structure, 25, 2017
|
|
2QPN
 
 | | GES-1 beta-lactamase | | Descriptor: | Beta-lactamase GES-1, SULFATE ION | | Authors: | Smith, C.A, Caccamo, M, Kantardjieff, K.A, Vakulenko, S. | | Deposit date: | 2007-07-24 | | Release date: | 2008-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of GES-1 at atomic resolution: insights into the evolution of carbapenamase activity in the class A extended-spectrum beta-lactamases.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5JFL
 
 | |
5TD6
 
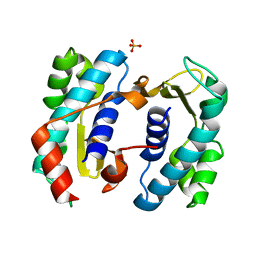 | | C. elegans FOG-3 BTG/Tob domain - H47N, C117A | | Descriptor: | FOG-3 protein, SULFATE ION | | Authors: | Aoki, S.T, Bingman, C.A, Wickens, M, Kimble, J.E. | | Deposit date: | 2016-09-17 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.034 Å) | | Cite: | An RNA-Binding Multimer Specifies Nematode Sperm Fate.
Cell Rep, 23, 2018
|
|
5TMK
 
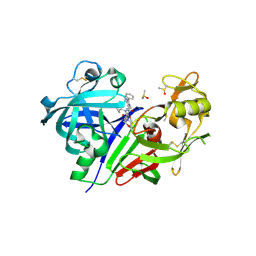 | | Optimization of 3,5-Disubstitued Piperidine: Discovery of Non-Peptide mimetics as an Orally Active Renin Inhibitor | | Descriptor: | 1-(4-methoxybutyl)-N-(2-methylpropyl)-N-[(3S,5R)-5-(morpholine-4-carbonyl)piperidin-3-yl]-5-phenyl-1H-pyrrole-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.C, Lane, W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Optimization of 3,5-Disubstitued Piperidine: Discovery of Non-Peptide mimetics as an Orally Active Renin Inhibitor
To be published
|
|
2QW7
 
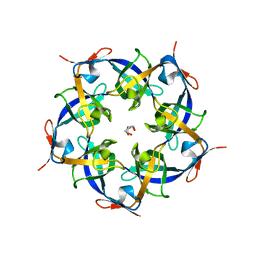 | | Carboxysome Subunit, CcmL | | Descriptor: | Carbon dioxide concentrating mechanism protein ccmL, GLYCEROL | | Authors: | Tanaka, S, Sawaya, M.R, Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 2007-08-09 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic-level models of the bacterial carboxysome shell.
Science, 319, 2008
|
|
2QI3
 
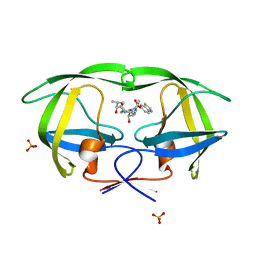 | | Crystal structure of protease inhibitor, MIT-2-AD94 in complex with wild type HIV-1 protease | | Descriptor: | (2S)-N-[(1S,2R)-3-{(1,3-BENZOTHIAZOL-6-YLSULFONYL)[(2S)-2-METHYLBUTYL]AMINO}-1-BENZYL-2-HYDROXYPROPYL]-2-HYDROXY-3-METHYLBUTANAMIDE, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
2PSV
 
 | |
2PTU
 
 | | Structure of NK cell receptor 2B4 (CD244) | | Descriptor: | Natural killer cell receptor 2B4 | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5TMG
 
 | | Optimization of 3,5-Disubstitued Piperidine: Discovery of Non-Peptide mimetics as an Orally Active Renin Inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(4-methoxybutyl)-N-(2-methylpropyl)-N-[(3S,5R)-5-(morpholine-4-carbonyl)piperidin-3-yl]-1-phenyl-1H-1,2,3-triazole-4-carboxamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.C, Lane, W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of 3,5-Disubstitued Piperidine: Discovery of Non-Peptide mimetics as an Orally Active Renin Inhibitor
To be published
|
|
2Q54
 
 | | Crystal structure of KB73 bound to HIV-1 protease | | Descriptor: | ACETATE ION, N~2~-ACETYL-N-[(1S,3S,4S)-4-({[(5S)-3-(3-ACETYLPHENYL)-2-OXO-1,3-OXAZOLIDIN-5-YL]CARBONYL}AMINO)-1-BENZYL-3-HYDROXY-5-PHENYLPENTYL]-L-VALINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-05-31 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design and Synthesis of HIV-1 Protease Inhibitors Incorporating Oxazolidinones as P2/P2' Ligands in Pseudosymmetric Dipeptide Isosteres.
J.Med.Chem., 50, 2007
|
|
3BUS
 
 | | Crystal Structure of RebM | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCoy, J.G, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of the rebeccamycin sugar 4'-O-methyltransferase RebM.
J.Biol.Chem., 283, 2008
|
|
3BZB
 
 | | Crystal structure of uncharacterized protein CMQ451C from the primitive red alga Cyanidioschyzon merolae | | Descriptor: | Uncharacterized protein | | Authors: | McCoy, J.G, Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2008-01-17 | | Release date: | 2008-01-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of uncharacterized protein CMQ451C from the primitive red alga Cyanidioschyzon merolae.
To be Published
|
|
2PTV
 
 | | Structure of NK cell receptor ligand CD48 | | Descriptor: | CD48 antigen | | Authors: | Deng, L, Velikovsky, C.A, Mariuzza, R.A. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of natural killer receptor 2B4 bound to CD48 reveals basis for heterophilic recognition in signaling lymphocyte activation molecule family.
Immunity, 27, 2007
|
|
2R2X
 
 | | Ricin A-chain (recombinant) complex with Urea | | Descriptor: | Ricin A chain, SULFATE ION, UREA | | Authors: | Carra, J.H, McHugh, C.A, Mulligan, S, Machiesky, L.M, Soares, A.S, Millard, C.B. | | Deposit date: | 2007-08-28 | | Release date: | 2007-11-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based identification of determinants of conformational and spectroscopic change at the ricin active site.
Bmc Struct.Biol., 7, 2007
|
|
5JH5
 
 | | Structural Basis for the Hierarchical Assembly of the Core of PRC1.1 | | Descriptor: | BCL-6 corepressor-like protein 1, Lysine-specific demethylase 2B, Polycomb group RING finger protein 1, ... | | Authors: | Wong, S.J, Taylor, A.B, Hart, P.J, Kim, C.A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1.
Structure, 24, 2016
|
|
5JFN
 
 | | Crystal structure of Rhodopseudomonas palustris propionaldehyde dehydrogenase with bound CoA and acylated Cys330 | | Descriptor: | Aldehyde dehydrogenase, COENZYME A, PENTAETHYLENE GLYCOL, ... | | Authors: | Zarzycki, J, Sutter, M, Kerfeld, C.A. | | Deposit date: | 2016-04-19 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In Vitro Characterization and Concerted Function of Three Core Enzymes of a Glycyl Radical Enzyme - Associated Bacterial Microcompartment.
Sci Rep, 7, 2017
|
|
3DCY
 
 | |
2Q55
 
 | | Crystal structure of KK44 bound to HIV-1 protease | | Descriptor: | (5S)-N-[(1S,2S,4S)-1-BENZYL-2-HYDROXY-4-{[(2S)-3-METHYL-2-(2-OXOTETRAHYDROPYRIMIDIN-1(2H)-YL)BUTANOYL]AMINO}-5-PHENYLPENTYL]-2-OXO-3-PHENYL-1,3-OXAZOLIDINE-5-CARBOXAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-05-31 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of HIV-1 Protease Inhibitors Incorporating Oxazolidinones as P2/P2' Ligands in Pseudosymmetric Dipeptide Isosteres.
J.Med.Chem., 50, 2007
|
|
2QI1
 
 | | Crystal structure of protease inhibitor, MIT-1-KK81 in complex with wild type HIV-1 protease | | Descriptor: | N-[(1S,2R)-1-BENZYL-2-HYDROXY-3-{[(3-METHOXYPHENYL)SULFONYL](2-THIENYLMETHYL)AMINO}PROPYL]-3,4-DIHYDROXYBENZAMIDE, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-07-03 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants.
J.Am.Chem.Soc., 130, 2008
|
|
3DI2
 
 | |
5JQD
 
 | | Antibody Fab Fragment | | Descriptor: | D80 Fab Fragment Heavy Chain, D80 Fab Fragment Light Chain | | Authors: | Zhang, Z, Prachanronarong, K, Gellatly, K, Marasco, W.A, Schiffer, C.A. | | Deposit date: | 2016-05-04 | | Release date: | 2017-11-08 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Structural Basis of an Influenza Hemagglutinin Stem-Directed Antibody Retaining the G6 Idiotype
To Be Published
|
|
