1D78
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1LHG
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BOROORNITHINE-OH | | Descriptor: | AC-(D)PHE-PRO-BOROHOMOORNITHINE-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
1LHF
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BORO-HOMOLYS-OH | | Descriptor: | AC-(D)PHE-PRO-BOROHOMOLYS-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
1LHC
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BOROARG-OH | | Descriptor: | AC-(D)PHE-PRO-BOROARG-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
1LHE
 
 | | HUMAN ALPHA-THROMBIN COMPLEXED WITH AC-(D)PHE-PRO-BORO-N-BUTYL-AMIDINO-GLYCINE-OH | | Descriptor: | AC-(D)PHE-PRO-BORO-N-BUTYL-AMIDINO-GLYCINE-OH, ALPHA-THROMBIN, HIRUDIN | | Authors: | Weber, P.C, Lee, S.L, Lewandowski, F.A, Schadt, M.C, Chang, C.H, Kettner, C.A. | | Deposit date: | 1994-12-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Kinetic and crystallographic studies of thrombin with Ac-(D)Phe-Pro-boroArg-OH and its lysine, amidine, homolysine, and ornithine analogs.
Biochemistry, 34, 1995
|
|
1LXC
 
 | | Crystal Structure of E. Coli Enoyl Reductase-NAD+ with a Bound Acrylamide Inhibitor | | Descriptor: | 3-(6-AMINOPYRIDIN-3-YL)-N-METHYL-N-[(1-METHYL-1H-INDOL-2-YL)METHYL]ACRYLAMIDE, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miller, W.H, Seefeld, M.A, Newlander, K.A, Uzinskas, I.N, Burgess, W.J, Heerding, D.A, Yuan, C.C.K, Head, M.S, Payne, D.J, Rittenhouse, S.F, Moore, T.D, Pearson, S.C, Dewolf, V, Berry, W.E, Keller, P.M, Polizzi, B.J, Qiu, X, Janson, C.A, Huffman, W.F. | | Deposit date: | 2002-06-05 | | Release date: | 2002-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of aminopyridine-based inhibitors of bacterial enoyl-ACP reductase (FabI).
J.Med.Chem., 45, 2002
|
|
1CQ3
 
 | | STRUCTURE OF A SOLUBLE SECRETED CHEMOKINE INHIBITOR, VCCI, FROM COWPOX VIRUS | | Descriptor: | VIRAL CHEMOKINE INHIBITOR | | Authors: | Carfi, A, Smith, C.A, Smolak, P.J, McGrew, J, Wiley, D.C. | | Deposit date: | 1999-08-05 | | Release date: | 1999-11-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a soluble secreted chemokine inhibitor vCCI (p35) from cowpox virus.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CQK
 
 | | CRYSTAL STRUCTURE OF THE CH3 DOMAIN FROM THE MAK33 ANTIBODY | | Descriptor: | CH3 DOMAIN OF MAK33 ANTIBODY | | Authors: | Thies, M.J, Mayer, J, Augustine, J.G, Frederick, C.A, Lilie, H, Buchner, J. | | Deposit date: | 1999-08-06 | | Release date: | 1999-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Folding and association of the antibody domain CH3: prolyl isomerization preceeds dimerization.
J.Mol.Biol., 293, 1999
|
|
1D13
 
 | | MOLECULAR STRUCTURE OF AN A-DNA DECAMER D(ACCGGCCGGT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*GP*CP*CP*GP*GP*T)-3') | | Authors: | Frederick, C.A, Quigley, G.J, Teng, M.-K, Coll, M, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular structure of an A-DNA decamer d(ACCGGCCGGT).
Eur.J.Biochem., 181, 1989
|
|
3OQ3
 
 | |
4QU3
 
 | | GES-2 ertapenem acyl-enzyme complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase GES-2, ... | | Authors: | Stewart, N.K, Smith, C.A, Frase, H, Black, D.J, Vakulenko, S.B. | | Deposit date: | 2014-07-10 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Kinetic and Structural Requirements for Carbapenemase Activity in GES-Type beta-Lactamases.
Biochemistry, 54, 2015
|
|
3SA6
 
 | | Crystal structure of wild-type HIV-1 protease in complex with AF71 | | Descriptor: | 3-hydroxy-N-[(2S,3R)-3-hydroxy-4-([(2S)-2-methylbutyl]{[5-(1,2-oxazol-5-yl)thiophen-2-yl]sulfonyl}amino)-1-phenylbutan-2-yl]benzamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
1MG4
 
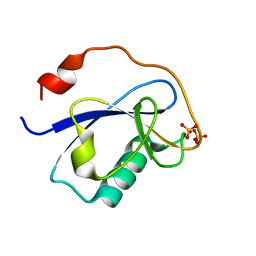 | | STRUCTURE OF N-TERMINAL DOUBLECORTIN DOMAIN FROM DCLK: WILD TYPE PROTEIN | | Descriptor: | DOUBLECORTIN-LIKE KINASE (N-TERMINAL DOMAIN), SULFATE ION | | Authors: | Kim, M.H, Cierpickil, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z. | | Deposit date: | 2002-08-14 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
3SA9
 
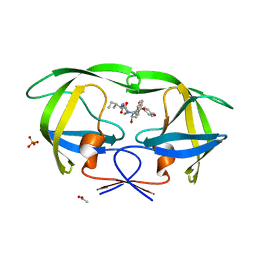 | | Crystal structure of Wild-type HIV-1 protease in complex With AF68 | | Descriptor: | ACETATE ION, N~2~-acetyl-N-[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]-L-isoleucinamide, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
1LUW
 
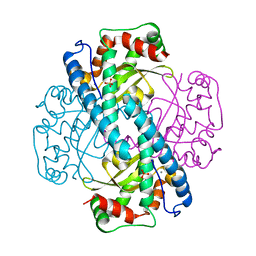 | | CATALYTIC AND STRUCTURAL EFFECTS OF AMINO-ACID SUBSTITUTION AT HIS 30 IN HUMAN MANGANESE SUPEROXIDE DISMUTASE: INSERTION OF VAL CGAMMA INTO THE SUBSTRATE ACCESS CHANNEL | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Superoxide dismutase [Mn] | | Authors: | Hearn, A.S, Stroupe, M.E, Ramilo, C.A, Luba, J.P, Cabelli, D.E, Tainer, J.A, Silverman, D.N. | | Deposit date: | 2002-05-23 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic and structural effects of amino acid substitution at histidine 30 in human manganese superoxide dismutase: insertion of valine C gamma into the substrate access channel
Biochemistry, 42, 2003
|
|
3SKH
 
 | |
4QX6
 
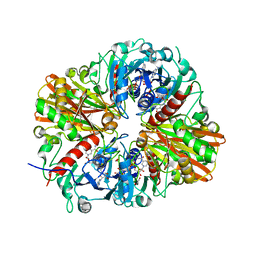 | | CRYSTAL STRUCTURE OF GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM STREPTOCOCCUS AGALACTIAE NEM316 at 2.46 ANGSTROM RESOLUTION | | Descriptor: | 1,2-ETHANEDIOL, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ayres, C.A, Schormann, N, Banerjee, S, Chattopadhyay, D. | | Deposit date: | 2014-07-18 | | Release date: | 2014-10-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of Streptococcus agalactiae glyceraldehyde-3-phosphate dehydrogenase holoenzyme reveals a novel surface.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4QY8
 
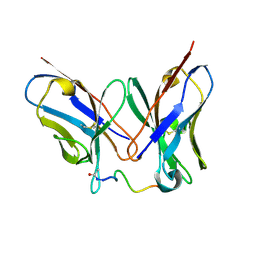 | | Crystal Structure of anti-MSP2 Fv fragment (mAb6D8) in complex with 3D7-MSP2 14-30 | | Descriptor: | Fv fragment(mAb6D8) heavy chain, Fv fragment(mAb6D8) light chain, Merozoite surface antigen 2 | | Authors: | Morales, R.A.V, MacRaild, C.A, Seow, J, Bankala, K, Drinkwater, N, McGowan, S, Rouet, R, Christ, D, Anders, R.F, Norton, R.S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.353 Å) | | Cite: | Structural basis for epitope masking and strain specificity of a conserved epitope in an intrinsically disordered malaria vaccine candidate.
Sci Rep, 5, 2015
|
|
3SA4
 
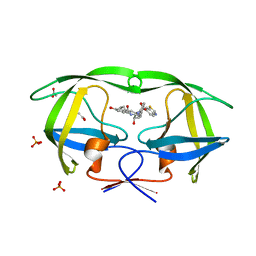 | | Crystal structure of wild-type HIV-1 protease in complex with AF72 | | Descriptor: | ACETATE ION, N-{(2S,3R)-4-[(1,3-benzothiazol-6-ylsulfonyl)(cyclohexylmethyl)amino]-3-hydroxy-1-phenylbutan-2-yl}-3-hydroxybenzamide, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SAC
 
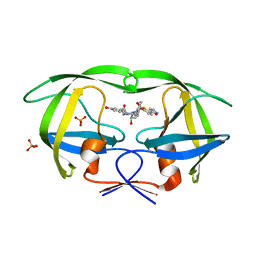 | | Crystal structure of wild-type HIV-1 protease in complex with AF80 | | Descriptor: | 3-hydroxy-N-{(2S,3R)-3-hydroxy-4-[(2-methylpropyl){[5-(1,2-oxazol-5-yl)thiophen-2-yl]sulfonyl}amino]-1-phenylbutan-2-yl}benzamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SA7
 
 | | Crystal structure of wild-type HIV-1 protease in complex with AF55 | | Descriptor: | N~2~-acetyl-N-[(2S,3R)-4-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-L-leucinamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SA8
 
 | | Crystal structure of wild-type HIV-1 protease in complex with KB83 | | Descriptor: | 3-hydroxy-N-[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]benzamide, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SKA
 
 | |
3SA3
 
 | | Crystal structure of wild-type HIV-1 protease in complex with AG23 | | Descriptor: | ACETATE ION, N~2~-acetyl-N-[(2S,3R)-4-{(1,3-benzothiazol-6-ylsulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]-L-isoleucinamide, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
3SAB
 
 | | Crystal structure of wild-type HIV-1 protease in complex with AF78 | | Descriptor: | 3-hydroxy-N-[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](3-phenylpropyl)amino}-1-phenylbutan-2-yl]benzamide, PHOSPHATE ION, Protease | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protease Inhibitors that protrude out from substrate envelope are more susceptible to developing drug resistance
To be Published
|
|
