7KFP
 
 | | Structure of human PARG complexed with PARG-119 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{[2-(1,3-dimethyl-2-oxo-6-sulfanylidene-1,2,3,6-tetrahydro-7H-purin-7-yl)ethyl]carbamoyl}methanesulfonamide, ... | | Authors: | Brosey, C.A, Bommagani, S, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG0
 
 | | Structure of human PARG complexed with PARG-131 | | Descriptor: | 1,2-ETHANEDIOL, 5-({4-[(1,3-dimethyl-2,6-dioxo-1,2,3,6-tetrahydro-7H-purin-7-yl)methyl]phenyl}methyl)pyrimidine-2,4,6(1H,3H,5H)-trione, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Arvai, A, Bommagani, S, Brosey, C.A, Jones, D.E, Warden, L.S, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-15 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7KG6
 
 | | Structure of human PARG complexed with PARG-322 | | Descriptor: | 1-{2-[(1,3-dimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)sulfanyl]ethyl}piperidine-4-carboxylic acid, Poly(ADP-ribose) glycohydrolase | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7L26
 
 | | HPK1 IN COMPLEX WITH COMPOUND 38 | | Descriptor: | 6-(2-fluoro-6-methylphenyl)-1-[4-(4-methylpiperazin-1-yl)phenyl]-1H-indazole-5-carbonitrile, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of Potent Reverse Indazole Inhibitors for HPK1.
Acs Med.Chem.Lett., 12, 2021
|
|
7KG8
 
 | | Structure of human PARG complexed with PARG-061 | | Descriptor: | 1,3-dimethyl-8-{[2-(morpholin-4-yl)-2-oxoethyl]sulfanyl}-6-sulfanylidene-1,3,6,7-tetrahydro-2H-purin-2-one, CACODYLATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Brosey, C.A, Balapiti-Modarage, L.P.F, Warden, L.S, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2020-10-16 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
7L24
 
 | | HPK1 IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 6-(2-fluoro-6-methoxyphenyl)-1-[4-(4-methylpiperazin-1-yl)phenyl]-1H-pyrazolo[4,3-c]pyridine, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Identification of Potent Reverse Indazole Inhibitors for HPK1.
Acs Med.Chem.Lett., 12, 2021
|
|
7L25
 
 | | HPK1 IN COMPLEX WITH COMPOUND 18 | | Descriptor: | 1,2-ETHANEDIOL, 6-(2-fluoro-6-methoxyphenyl)-1-[6-(4-methylpiperazin-1-yl)pyridin-2-yl]-1H-pyrazolo[4,3-c]pyridine, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Lesburg, C.A. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of Potent Reverse Indazole Inhibitors for HPK1.
Acs Med.Chem.Lett., 12, 2021
|
|
7KYL
 
 | | Powassan virus Envelope protein DIII in complex with neutralizing Fab POWV-80 | | Descriptor: | CHLORIDE ION, Envelope protein domain III, POWV-80 Fab heavy chain, ... | | Authors: | Errico, J.M, Nelson, C.A, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broadly neutralizing monoclonal antibodies protect against multiple tick-borne flaviviruses.
J.Exp.Med., 218, 2021
|
|
4APU
 
 | | PR X-Ray structures in agonist conformations reveal two different mechanisms for partial agonism in 11beta-substituted steroids | | Descriptor: | (8S,11R,13S,14S,16S,17S)-17-cyclopropylcarbonyl-16-ethenyl-13-methyl-11-(4-pyridin-3-ylphenyl)-2,6,7,8,11,12,14,15,16,17-decahydro-1H-cyclopenta[a]phenanthren-3-one, 2-CHLORO-N-[[4-(3,5-DIMETHYLISOXAZOL-4-YL)PHENYL]METHYL]-1,4-DIMETHYL-1H-PYRAZOLE-4-SULFONAMIDE, PROGESTERONE RECEPTOR, ... | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Bosch, R, Vu-Pham, D, McGuire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of progesterone receptor ligand binding domain in its agonist state reveal differing mechanisms for mixed profiles of 11 beta-substituted steroids.
J. Biol. Chem., 287, 2012
|
|
7JP2
 
 | | Crystal structure of TP0037 from Treponema pallidum, a D-lactate dehydrogenase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-lactate dehydrogenase | | Authors: | Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2020-08-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Biophysical and Biochemical Characterization of TP0037, a d-Lactate Dehydrogenase, Supports an Acetogenic Energy Conservation Pathway in Treponema pallidum.
Mbio, 11, 2020
|
|
7K8M
 
 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C102 Fab Heavy Chain, C102 Fab Light Chain, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8O
 
 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C002 | | Descriptor: | C002 Fab Heavy Chain, C002 Fab Light Chain, GLYCEROL, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
4ADY
 
 | | Crystal structure of 26S proteasome subunit Rpn2 | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN2 | | Authors: | Kulkarni, K, He, J, Da Fonseca, P.C.A, Krutauz, D, Glickman, M.H, Barford, D, Morris, E.P. | | Deposit date: | 2012-01-04 | | Release date: | 2012-03-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Structure of the 26S Proteasome Subunit Rpn2 Reveals its Pc Repeat Domain as a Closed Toroid of Two Concentric Alpha-Helical Rings
Structure, 20, 2012
|
|
7KNN
 
 | |
6QBS
 
 | | The Alkyne Moiety as a Latent Electrophile in Irreversible Covalent Small Molecule Inhibitors of Cathepsin K | | Descriptor: | (2~{S})-4-methyl-~{N}-prop-2-enyl-2-[[(1~{S})-2,2,2-tris(fluoranyl)-1-[4-(4-methylsulfonylphenyl)phenyl]ethyl]amino]pentanamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Mons, E, Jansen, I.D.C, Loboda, J, van Doodewaerd, B.R, Verdoes, M, van Boeckel, C.A.A, van Veelen, P.A, Turk, B, Turk, D, Hermans, J, Ovaa, H. | | Deposit date: | 2018-12-21 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Alkyne Moiety as a Latent Electrophile in Irreversible Covalent Small Molecule Inhibitors of Cathepsin K.
J. Am. Chem. Soc., 141, 2019
|
|
7JJ9
 
 | | Crystal structure of Zn(II)-bound AdcA from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, ZINC ION, Zinc-binding lipoprotein AdcA | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
7JJ8
 
 | | Crystal structure of the Zn(II)-bound ZnuA-like domain of Streptococcus pneumoniae AdcA | | Descriptor: | ZINC ION, Zinc-binding lipoprotein AdcA | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
7JIS
 
 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 2F | | Descriptor: | (3S)-3-benzyl-5-[9-ethyl-8-(2-methylpyrimidin-5-yl)-9H-purin-6-yl]-3-methyl-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2020-07-23 | | Release date: | 2020-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Optimization of Versatile Oxindoles as Selective PI3K delta Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
7JJB
 
 | | Crystal structure of Zn(II)-bound ZinT-like domain of Streptococcus pneumoniae AdcA | | Descriptor: | MAGNESIUM ION, SODIUM ION, ZINC ION, ... | | Authors: | Luo, Z, More, J.R, Kobe, B, McDevitt, C.A. | | Deposit date: | 2020-07-24 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | A Trap-Door Mechanism for Zinc Acquisition by Streptococcus pneumoniae AdcA.
Mbio, 12, 2021
|
|
1VK0
 
 | | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450 | | Descriptor: | hypothetical protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-04-12 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structure of Gene Product from Arabidopsis Thaliana At5g06450
To be published
|
|
7JIU
 
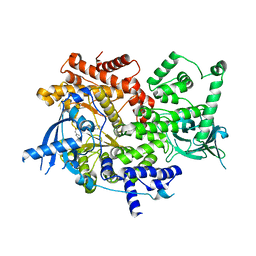 | | HUMAN PI3KDELTA IN COMPLEX WITH COMPOUND 2F | | Descriptor: | (3S)-3-benzyl-5-[9-ethyl-8-(2-methylpyrimidin-5-yl)-9H-purin-6-yl]-3-methyl-1,3-dihydro-2H-indol-2-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Lesburg, C.A, Augustin, M. | | Deposit date: | 2020-07-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Optimization of Versatile Oxindoles as Selective PI3K delta Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6OWZ
 
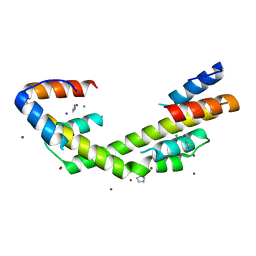 | | Spy H96L:Im7 L19pI-Phe complex; multiple anomalous datasets contained herein for element identification | | Descriptor: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | Authors: | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | Deposit date: | 2019-05-12 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4BOY
 
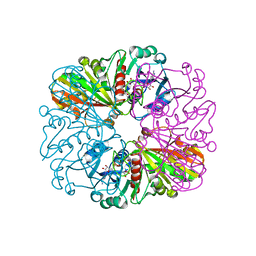 | |
6QJB
 
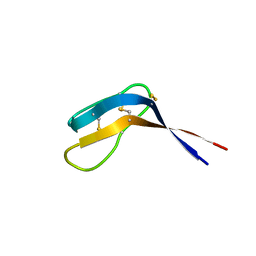 | | Truncated Evasin-3 (tEv3 17-56) | | Descriptor: | Evasin-3 | | Authors: | Denisov, S.S, Ippel, J.H, Heinzman, A.C.A, Koenen, R.R, Ortega-Gomez, A, Soehnlein, O, Hackeng, T.M, Dijkgraaf, I. | | Deposit date: | 2019-01-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Tick saliva protein Evasin-3 modulates chemotaxis by disrupting CXCL8 interactions with glycosaminoglycans and CXCR2.
J.Biol.Chem., 294, 2019
|
|
4B7M
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
