1FH5
 
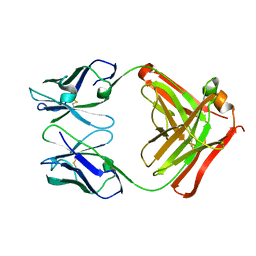 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MONOCLONAL ANTIBODY MAK33 | | Descriptor: | MONOCLONAL ANTIBODY MAK33 | | Authors: | Augustine, J.G, de la Calle, A, Knarr, G, Buchner, J, Frederick, C.A. | | Deposit date: | 2000-07-31 | | Release date: | 2000-09-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of the fab fragment of the monoclonal antibody MAK33. Implications for folding and interaction with the chaperone bip.
J.Biol.Chem., 276, 2001
|
|
4U2C
 
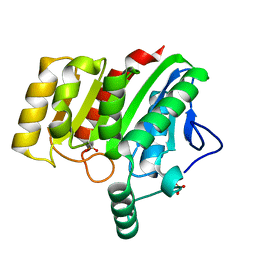 | | Crystal structure of dienelactone hydrolase A-6 variant (S7T, A24V, Q35H, F38L, Q110L, C123S, Y145C, E199G and S208G) at 1.95 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2D
 
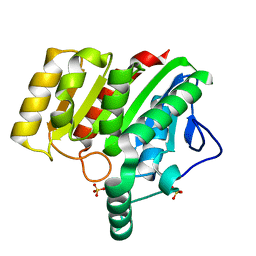 | | Crystal structure of dienelactone hydrolase S-2 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G and G211D) at 1.67 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2G
 
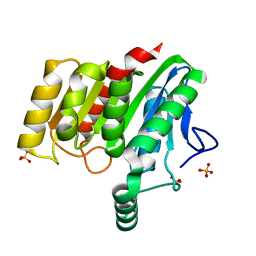 | | Crystal structure of dienelactone hydrolase B-4 variant (Q35H, F38L, Y64H, Q76L, Q110L, C123S, Y137C, A141V, Y145C, N154D, E199G, S208G, G211D, S233G and 237Q) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
1FNG
 
 | | HISTOCOMPATIBILITY ANTIGEN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (MHC CLASS II I-EK, ALPHA CHAIN), ... | | Authors: | Miley, M.J, Nelson, C.A, Fremont, D.H. | | Deposit date: | 2000-08-21 | | Release date: | 2001-03-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional consequences of altering a peptide MHC anchor residue.
J.Immunol., 166, 2001
|
|
1FQL
 
 | | X-RAY CRYSTAL STRUCTURE OF ZINC-BOUND F95M/W97V CARBONIC ANHYDRASE (CAII) VARIANT | | Descriptor: | CARBONIC ANHYDRASE, MERCURY (II) ION, ZINC ION | | Authors: | Cox, J.D, Hunt, J.A, Compher, K.M, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2000-09-05 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural influence of hydrophobic core residues on metal binding and specificity in carbonic anhydrase II.
Biochemistry, 39, 2000
|
|
4YU7
 
 | | Crystal structure of Piratoxin I (PrTX-I) complexed to caffeic acid | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Basic phospholipase A2 homolog piratoxin-1, CAFFEIC ACID, ... | | Authors: | Fernandes, C.A.H, Fontes, M.R.M. | | Deposit date: | 2015-03-18 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Structural Basis for the Inhibition of a Phospholipase A2-Like Toxin by Caffeic and Aristolochic Acids.
Plos One, 10, 2015
|
|
1D79
 
 | | HIGH RESOLUTION REFINEMENT OF THE HEXAGONAL A-DNA OCTAMER D(GTGTACAC) AT 1.4 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*AP*CP*AP*C)-3') | | Authors: | Thota, N, Li, X.H, Bingman, C.A, Sundaralingam, M. | | Deposit date: | 1992-06-12 | | Release date: | 1993-04-15 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution refinement of the hexagonal A-DNA octamer d(GTGTACAC) at 1.4 A.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4YWK
 
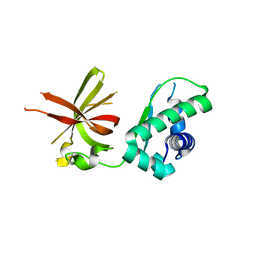 | |
1D12
 
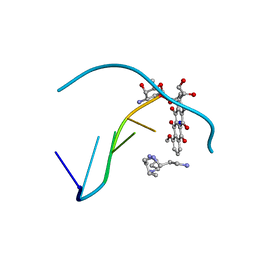 | | STRUCTURAL COMPARISON OF ANTICANCER DRUG-DNA COMPLEXES. ADRIAMYCIN AND DAUNOMYCIN | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), DOXORUBICIN, SODIUM ION, ... | | Authors: | Frederick, C.A, Williams, L.D, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin.
Biochemistry, 29, 1990
|
|
1CYJ
 
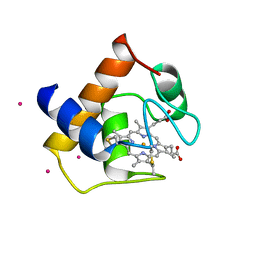 | | CYTOCHROME C6 | | Descriptor: | CADMIUM ION, CYTOCHROME C6, HEME C | | Authors: | Kerfeld, C.A, Yeates, T.O. | | Deposit date: | 1995-05-09 | | Release date: | 1996-01-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of chloroplast cytochrome c6 at 1.9 A resolution: evidence for functional oligomerization.
J.Mol.Biol., 250, 1995
|
|
5A2S
 
 | | Potent, selective and CNS-penetrant tetrasubstituted cyclopropane class IIa histone deacetylase (HDAC) inhibitors | | Descriptor: | (1S,2S,3S)-1-fluoranyl-2-[4-(5-fluoranylpyrimidin-2-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, SODIUM ION, ... | | Authors: | Luckhurst, C.A, Breccia, P, Stott, A.J, Aziz, O, Birch, H, Burli, R.W, Hughes, S, Jarvis, R.E, Lamers, M, Leonard, P, Matthews, K.L, McAllister, G, Pollack, S, Saville-Stones, E, Wishart, G, Yates, D, Dominguez, C. | | Deposit date: | 2015-05-22 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Potent, Selective, and Cns-Penetrant Tetrasubstituted Cyclopropane Class Iia Histone Deacetylase (Hdac) Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
1DNF
 
 | | EFFECTS OF 5-FLUOROURACIL/GUANINE WOBBLE BASE PAIRS IN Z-DNA. MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGFG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(UFP)P*G)-3'), MAGNESIUM ION | | Authors: | Coll, M, Saal, D, Frederick, C.A, Aymami, J, Rich, A, Wang, A.H.-J. | | Deposit date: | 1988-12-12 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Effects of 5-fluorouracil/guanine wobble base pairs in Z-DNA: molecular and crystal structure of d(CGCGFG).
Nucleic Acids Res., 17, 1989
|
|
4YWL
 
 | |
4U3Q
 
 | |
4U6Q
 
 | | CtBP1 bound to inhibitor 2-(hydroxyimino)-3-phenylpropanoic acid | | Descriptor: | (2E)-2-(hydroxyimino)-3-phenylpropanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C-terminal-binding protein 1, ... | | Authors: | Hilbert, B.J, Morris, B.L, Ellis, K.C, Paulsen, J.L, Schiffer, C.A, Grossman, S.R, Royer Jr, W.E. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Guided Design of a High Affinity Inhibitor to Human CtBP.
Acs Chem.Biol., 10, 2015
|
|
1G3O
 
 | | CRYSTAL STRUCTURE OF V19E MUTANT OF FERREDOXIN I | | Descriptor: | 7FE FERREDOXIN I, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER | | Authors: | Stout, C.D, Burgess, B.K, Bonagura, C.A, Jung, Y.S. | | Deposit date: | 2000-10-24 | | Release date: | 2000-11-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Azotobacter vinelandii ferredoxin I: a sequence and structure comparison approach to alteration of [4Fe-4S]2+/+ reduction potential
J.Biol.Chem., 277, 2002
|
|
1G54
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3,4,5,6-PENTAFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2,3,4,5,6-PENTAFLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-30 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Contribution of Flourine to Protein-ligand Affinity in the Binding of Flouroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
4V5H
 
 | | E.Coli 70s Ribosome Stalled During Translation Of Tnac Leader Peptide. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Seidelt, B, Innis, C.A, Wilson, D.N, Gartmann, M, Armache, J, Villa, E, Trabuco, L.G, Becker, T, Mielke, T, Schulten, K, Steitz, T.A, Beckmann, R. | | Deposit date: | 2009-10-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural insight into nascent polypeptide chain-mediated translational stalling.
Science, 326, 2009
|
|
1FZI
 
 | | METHANE MONOOXYGENASE HYDROXYLASE, FORM I PRESSURIZED WITH XENON GAS | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE COMPONENT A, ALPHA CHAIN, ... | | Authors: | Whittington, D.A, Rosenzweig, A.C, Frederick, C.A, Lippard, S.J. | | Deposit date: | 2000-10-03 | | Release date: | 2001-04-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Xenon and halogenated alkanes track putative substrate binding cavities in the soluble methane monooxygenase hydroxylase.
Biochemistry, 40, 2001
|
|
4U2B
 
 | |
4U2E
 
 | | Crystal structure of dienelactone hydrolase S-3 variant (Q35H, F38L, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G, G211D and K234N) at 1.70 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
4U2F
 
 | | Crystal structure of dienelactone hydrolase B-1 variant (Q35H, F38L, Y64H, Q110L, C123S, Y137C, Y145C, N154D, E199G, S208G and G211D) at 1.80 A resolution | | Descriptor: | Carboxymethylenebutenolidase, SULFATE ION | | Authors: | Porter, J.L, Collyer, C.A, Ollis, D.L. | | Deposit date: | 2014-07-16 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed evolution of new and improved enzyme functions using an evolutionary intermediate and multidirectional search.
Acs Chem.Biol., 10, 2015
|
|
1F8F
 
 | | CRYSTAL STRUCTURE OF BENZYL ALCOHOL DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS | | Descriptor: | BENZYL ALCOHOL DEHYDROGENASE, ETHANOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Beauchamp, J.C, Gillooly, D, Warwicker, J, Fewson, C.A, Lapthorn, A.J. | | Deposit date: | 2000-06-30 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Benzyl Alcohol Dehydrogenase from Acinetobacter calcoaceticus
To be Published
|
|
4ZJ8
 
 | | Structures of the human OX1 orexin receptor bound to selective and dual antagonists | | Descriptor: | OLEIC ACID, [(7R)-4-(5-chloro-1,3-benzoxazol-2-yl)-7-methyl-1,4-diazepan-1-yl][5-methyl-2-(2H-1,2,3-triazol-2-yl)phenyl]methanone, human OX1R fusion protein to P.abysii glycogen synthase | | Authors: | Yin, J, Brautigam, C.A, Shao, Z, Clark, L, Harrell, C.M, Gotter, A.L, Coleman, P, Renger, J.J, Rosenbaum, D.M. | | Deposit date: | 2015-04-29 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structure and ligand-binding mechanism of the human OX1 and OX2 orexin receptors.
Nat.Struct.Mol.Biol., 23, 2016
|
|
