4HX6
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 | | Descriptor: | ACETATE ION, Oxidoreductase, SULFATE ION | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-09 | | Release date: | 2012-11-28 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
1MBZ
 
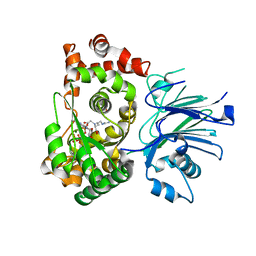 | | BETA-LACTAM SYNTHETASE WITH TRAPPED INTERMEDIATE | | Descriptor: | ARGININE-N-METHYLCARBONYL PHOSPHORIC ACID 5'-ADENOSINE ESTER, BETA-LACTAM SYNTHETASE, GLYCEROL, ... | | Authors: | Miller, M.T, Bachmann, B.O, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2002-08-04 | | Release date: | 2002-10-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The catalytic cycle of beta -lactam synthetase observed by x-ray crystallographic snapshots
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4HPL
 
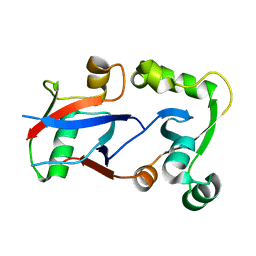 | | PCGF1 Ub fold (RAWUL)/BCOR PUFD Complex | | Descriptor: | BCL-6 corepressor, Polycomb group RING finger protein 1 | | Authors: | Junco, S.E, Wang, R, Gaipa, J, Taylor, A.B, Gearhart, M.D, Bardwell, V.J, Hart, P.J, Kim, C.A. | | Deposit date: | 2012-10-24 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Polycomb Group Protein PCGF1 in Complex with BCOR Reveals Basis for Binding Selectivity of PCGF Homologs.
Structure, 21, 2013
|
|
4HT5
 
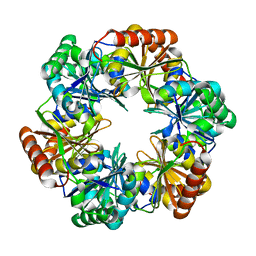 | | CO2 concentrating mechanism protein P, CcmP form 1 | | Descriptor: | CO2 concentrating mechanism protein P | | Authors: | Cai, F, Sutter, M, Kerfeld, C.A. | | Deposit date: | 2012-10-31 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment.
J.Biol.Chem., 288, 2013
|
|
1MG7
 
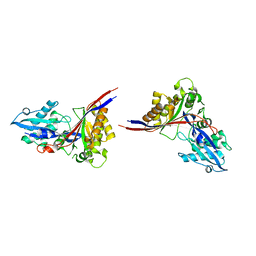 | | Crystal Structure of xol-1 | | Descriptor: | early switch protein xol-1 2.2k splice form | | Authors: | Luz, J.G, Hassig, C.A, Godzik, A, Meyer, B.J, Wilson, I.A. | | Deposit date: | 2002-08-14 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | XOL-1, primary determinant of sexual fate in C. elegans, is a GHMP kinase family member and a structural prototype for a class of developmental regulators
Genes Dev., 17, 2003
|
|
4HVM
 
 | | Crystal structure of tallysomycin biosynthesis protein TlmII | | Descriptor: | SULFATE ION, TlmII | | Authors: | Chang, C, Bigelow, L, Bearden, J, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of tallysomycin biosynthesis protein TlmII
To be Published
|
|
4HZP
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP Acyltransferase/Decarboxylase, CHLORIDE ION, COENZYME A, ... | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
2OZA
 
 | | Structure of p38alpha complex | | Descriptor: | MAP kinase-activated protein kinase 2, Mitogen-activated protein kinase 14 | | Authors: | White, A, Pargellis, C.A, Studts, J.M, Werneburg, B.G, Farmer II, B.T. | | Deposit date: | 2007-02-25 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of MAPK-activated protein kinase 2:p38 assembly
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2GRM
 
 | | Crystal structure of PrgX/iCF10 complex | | Descriptor: | PrgX, peptide | | Authors: | Shi, K, Kozlowicz, B.K, Gu, Z.Y, Ohlendorf, D.H, Earhart, C.A, Dunny, G.M. | | Deposit date: | 2006-04-24 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides.
Mol.Microbiol., 62, 2006
|
|
4HZN
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional Methylmalonyl-CoA:ACP Acyltransferase/Decarboxylase, GLYCEROL, ... | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
3TYV
 
 | |
3U64
 
 | | The Crystal Structure of Tat-T (Tp0956) | | Descriptor: | Protein TP_0956, SULFATE ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural, Bioinformatic, and In Vivo Analyses of Two Treponema pallidum Lipoproteins Reveal a Unique TRAP Transporter.
J.Mol.Biol., 416, 2012
|
|
2GNX
 
 | | X-ray structure of a hypothetical protein from Mouse Mm.209172 | | Descriptor: | hypothetical protein | | Authors: | Phillips Jr, G.N, McCoy, J.G, Bitto, E, Wesenberg, G.E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-11 | | Release date: | 2006-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | X-ray structure of a hypothetical protein from Mouse Mm.209172
To be Published
|
|
2MBQ
 
 | |
2GO3
 
 | | Crystal structure of Aquifex aeolicus LpxC complexed with imidazole. | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Li, X, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2006-04-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic Inferences from the Binding of Ligands to LpxC, a Metal-Dependent Deacetylase
Biochemistry, 45, 2006
|
|
2MBO
 
 | |
4HZO
 
 | | The Structure of the Bifunctional Acetyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double Hot Dog Fold | | Descriptor: | Bifunctional methylmalonyl-CoA:ACP acyltransferase/decarboxylase, CHLORIDE ION, COENZYME A | | Authors: | Lohman, J.R, Bingman, C.A, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-15 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure of the Bifunctional Acyltransferase/Decarboxylase LnmK from the Leinamycin Biosynthetic Pathway Revealing Novel Activity for a Double-Hot-Dog Fold.
Biochemistry, 52, 2013
|
|
2LZZ
 
 | | Solution structure of a mutant of the triheme cytochrome PpcA from Geobacter sulfurreducens sheds light on the role of the conserved aromatic residue F15 | | Descriptor: | Cytochrome c, 3 heme-binding sites, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dantas, J.M, Morgado, L, Turner, D.L, Salgueiro, C.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-01-30 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a mutant of the triheme cytochrome PpcA from Geobacter sulfurreducens sheds light on the role of the conserved aromatic residue F15.
Biochim.Biophys.Acta, 1827, 2013
|
|
2MFY
 
 | | Non-reducible analogues of alpha-conotoxin Vc1.1: [2,8]-trans dicarba Vc1.1 | | Descriptor: | Alpha-conotoxin Vc1A | | Authors: | Robinson, S.D, Macraild, C.A, Van Lierop, B.J, Robinson, A.J, Norton, R.S. | | Deposit date: | 2013-10-24 | | Release date: | 2013-12-18 | | Method: | SOLUTION NMR | | Cite: | Dicarba alpha-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors.
Acs Chem.Biol., 8, 2013
|
|
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
2PC4
 
 | | Crystal structure of fructose-bisphosphate aldolase from Plasmodium falciparum in complex with TRAP-tail determined at 2.4 angstrom resolution | | Descriptor: | Fructose-bisphosphate aldolase, PbTRAP | | Authors: | Bosch, J, Buscaglia, C.A, Krumm, B, Cardozo, T, Nussenzweig, V, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3ZDG
 
 | | Crystal Structure of Ls-AChBP complexed with carbamoylcholine analogue 3-(dimethylamino)butyl dimethylcarbamate (DMABC) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(dimethylamino)butyl dimethylcarbamate, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Ussing, C.A, Hansen, C.P, Petersen, J.G, Jensen, A.A, Rohde, L.A.H, Ahring, P.K, Nielsen, E.O, Kastrup, J.S, Gajhede, M, Frolund, B, Balle, T. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Synthesis, Pharmacology, and Biostructural Characterization of Novel Alpha(4)Beta(2) Nicotinic Acetylcholine Receptor Agonists.
J.Med.Chem., 56, 2013
|
|
3ZDH
 
 | | Crystal structure of Ls-AChBP complexed with carbamoylcholine analogue N,N-dimethyl-4-(1-methyl-1H-imidazol-2-yloxy)butan-2-amine | | Descriptor: | (2R)-N,N-dimethyl-4-(1-methylimidazol-2-yl)oxy-butan-2-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Ussing, C.A, Hansen, C.P, Petersen, J.G, Jensen, A.A, Rohde, L.A.H, Ahring, P.K, Nielsen, E.O, Kastrup, J.S, Gajhede, M, Frolund, B, Balle, T. | | Deposit date: | 2012-11-26 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Synthesis, Pharmacology, and Biostructural Characterization of Novel Alpha(4)Beta(2) Nicotinic Acetylcholine Receptor Agonists.
J.Med.Chem., 56, 2013
|
|
2K9E
 
 | |
2PSU
 
 | | Crystal Structure of wild type HIV-1 protease in complex with CARB-AD37 | | Descriptor: | ACETATE ION, N-[(1S,2R)-1-BENZYL-3-{(CYCLOPROPYLMETHYL)[(3-METHOXYPHENYL)SULFONYL]AMINO}-2-HYDROXYPROPYL]-N'-METHYLSUCCINAMIDE, PHOSPHATE ION, ... | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2007-05-07 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of Mutation-resistant HIV Protease Inhibitors with the Substrate Envelope Hypothesis.
Chem.Biol.Drug Des., 69, 2007
|
|
