3TSI
 
 | | Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) stalk domain | | Descriptor: | Hemagglutinin-neuraminidase | | Authors: | Bose, S, Welch, B.D, Kors, C.A, Yuan, P, Jardetzky, T.S, Lamb, R.A. | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion.
J.Virol., 85, 2011
|
|
6UJS
 
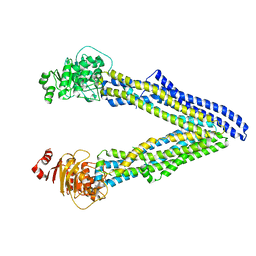 | | P-glycoprotein mutant-F728A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
1NL6
 
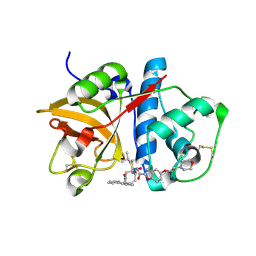 | |
6UJN
 
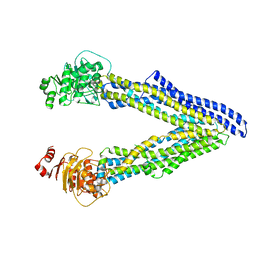 | | P-glycoprotein mutant-C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
4E73
 
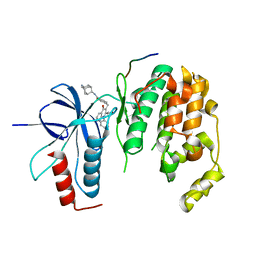 | | Crystal structure of JNK1beta-JIP in complex with an azaquinolone inhbitor | | Descriptor: | C-Jun-amino-terminal kinase-interacting protein 1, Mitogen-activated protein kinase 8, methyl 3-(4-{[(1R,2S,3S,5S,7s)-5-aminotricyclo[3.3.1.1~3,7~]dec-2-yl]carbamoyl}benzyl)-4-oxo-1-phenyl-1,4-dihydro-1,8-naphthyridine-2-carboxylate | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2012-03-16 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Identification of an Adamantyl Azaquinolone JNK Selective Inhibitor.
ACS Med Chem Lett, 3, 2012
|
|
6UKY
 
 | | STING C-terminal Domain Complexed with Non-cyclic Dinucleotide Compound 12 | | Descriptor: | 4-(6-{3-[2-(3-carboxypropanoyl)-6-methoxy-1-benzothiophen-4-yl]propyl}-5-methoxy-1-benzothiophen-2-yl)-4-oxobutanoic acid, fusion protein of Ubiquitin-like protein SMT3 and Stimulator of interferon protein c-terminal domain | | Authors: | Lesburg, C.A. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An orally available non-nucleotide STING agonist with antitumor activity.
Science, 369, 2020
|
|
2LQR
 
 | | NMR structure of Ig3 domain of palladin | | Descriptor: | Palladin | | Authors: | Beck, M.R, Dixon IV, R.D.S, Otey, C.A, Campbell, S.L, Murphy, G.S. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Function of Palladin's Actin Binding Domain.
J.Mol.Biol., 425, 2013
|
|
1NE8
 
 | | YDCE protein from Bacillus subtilis | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETIC ACID, conserved hypothetical protein YDCE | | Authors: | Gogos, A, Mu, H, Bahna, F, Gomez, C.A, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-12-10 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of YdcE protein from Bacillus subtilis
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
6UD0
 
 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | Descriptor: | Tumor susceptibility gene 101 protein, Ubiquitin | | Authors: | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | Deposit date: | 2019-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
3U27
 
 | | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b | | Descriptor: | CALCIUM ION, GLYCEROL, Microcompartments protein, ... | | Authors: | Wu, R, Gu, M, Kerfeld, C.A, Salmeen, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-10-01 | | Release date: | 2012-02-08 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Crystal structure of ethanolamine utilization protein EutL from Leptotrichia buccalis C-1013-b
To be Published
|
|
3KUY
 
 | |
2G83
 
 | | Structure of activated G-alpha-i1 bound to a nucleotide-state-selective peptide: Minimal determinants for recognizing the active form of a G protein alpha subunit | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Johnston, C.A, Ramer, J.K, Blaesius, R, Kuhlman, B, Arshavsky, V.Y, Siderovski, D.P. | | Deposit date: | 2006-03-01 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Minimal Determinants for Binding Activated Galpha from the Structure of a Galpha(i1)-Peptide Dimer.
Biochemistry, 45, 2006
|
|
2LDL
 
 | | Solution NMR Structure of the HIV-1 Exon Splicing Silencer 3 | | Descriptor: | RNA (27-MER) | | Authors: | Mishler, C, Levengood, J.D, Johnson, C.A, Rajan, P, Znosko, B.M. | | Deposit date: | 2011-05-27 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the HIV-1 Exon Splicing Silencer 3.
J.Mol.Biol., 415, 2012
|
|
1M1Z
 
 | |
4IFZ
 
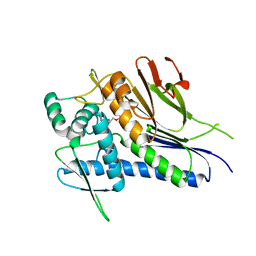 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, Mn(II)-AMP product bound form | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9012 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
1LX6
 
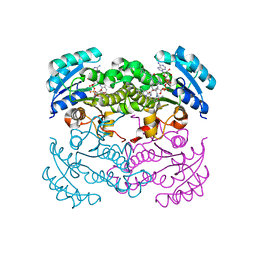 | |
6B4H
 
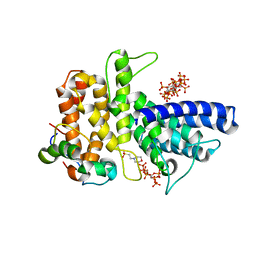 | | Crystal structure of Chaetomium thermophilum Gle1 CTD-Nup42 GBM-IP6 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, INOSITOL HEXAKISPHOSPHATE, Nucleoporin AMO1, ... | | Authors: | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
3V5Z
 
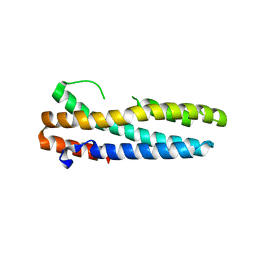 | | Structure of FBXL5 hemerythrin domain, C2 cell, grown anaerobically | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1847 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
1LUV
 
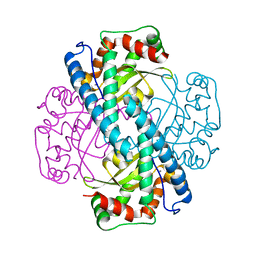 | | CATALYTIC AND STRUCTURAL EFFECTS OF AMINO-ACID SUBSTITUTION AT HIS 30 IN HUMAN MANGANESE SUPEROXIDE DISMUTASE: INSERTION OF VAL CGAMMA INTO THE SUBSTRATE ACCESS CHANNEL | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn] | | Authors: | Hearn, A.S, Stroupe, M.E, Ramilo, C.A, Luba, J.P, Cabelli, D.E, Tainer, J.A, Silverman, D.N. | | Deposit date: | 2002-05-23 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catalytic and structural effects of amino acid substitution at histidine 30 in human manganese superoxide dismutase: insertion of valine C gamma into the substrate access channel
Biochemistry, 42, 2003
|
|
6V6G
 
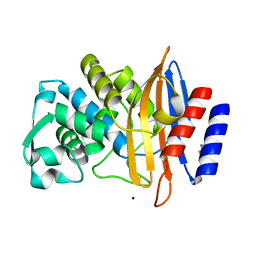 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
4IJZ
 
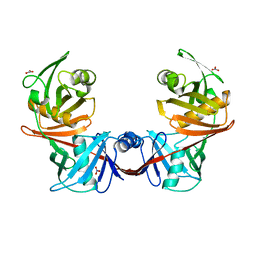 | | Crystal structure of diaminopimelate epimerase from Escherichia coli | | Descriptor: | Diaminopimelate epimerase, NITRATE ION | | Authors: | Hor, L, Dobson, R.C.J, Hutton, C.A, Perugini, M.A. | | Deposit date: | 2012-12-24 | | Release date: | 2013-02-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Dimerization of bacterial diaminopimelate epimerase is essential for catalysis
J.Biol.Chem., 288, 2013
|
|
6V6P
 
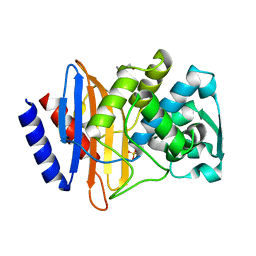 | | Crystal structure of CTX-M-14 E166A/D240G beta-lactamase | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6B4E
 
 | | Crystal structure of Saccharomyces cerevisiae Gle1 CTD-Nup42 GBM complex | | Descriptor: | 1,2-ETHANEDIOL, Nucleoporin GLE1, Nucleoporin NUP42, ... | | Authors: | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
2LFV
 
 | | Solution Structure of the SPOR domain from E. coli DamX | | Descriptor: | Protein damX | | Authors: | Williams, K.B, Arends, S.J.R, Popham, D.L, Fowler, C.A, Weiss, D.S. | | Deposit date: | 2011-07-15 | | Release date: | 2012-07-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of the Peptidoglycan-Binding SPOR Domain from Escherichia coli DamX: Insights into Septal Localization.
Biochemistry, 52, 2013
|
|
1MJD
 
 | | Structure of N-terminal domain of human doublecortin | | Descriptor: | DOUBLECORTIN | | Authors: | Kim, M.H, Cierpicki, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z.S. | | Deposit date: | 2002-08-27 | | Release date: | 2003-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
