1KIB
 
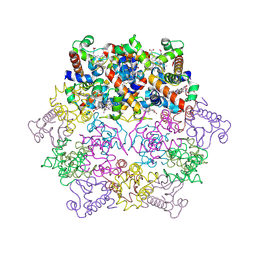 | | cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in the form of an oblate shell | | Descriptor: | HEME C, cytochrome c6 | | Authors: | Kerfeld, C.A, Sawaya, M.R, Krogmann, D, Yeates, T.O. | | Deposit date: | 2001-12-03 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of cytochrome c6 from Arthrospira maxima: an assembly of 24 subunits in a nearly symmetric shell.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
7V0J
 
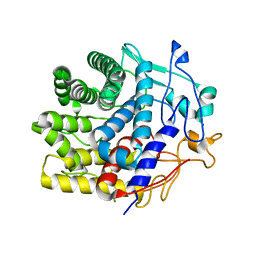 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellobiose product | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7UNP
 
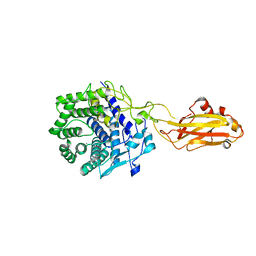 | | Crystal structure of the CelR catalytic domain and CBM3c | | Descriptor: | CALCIUM ION, Glucanase | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7V0I
 
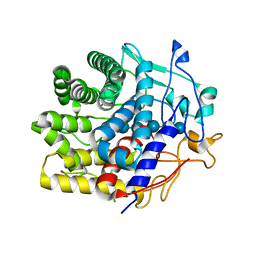 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellohexaose substrate | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
5KOT
 
 | | Discovery of TAK-272: A Novel, Potent and Orally Active Renin In-hibitor | | Descriptor: | 1-(4-methoxybutyl)-~{N}-(2-methylpropyl)-~{N}-[(3~{S},5~{R})-5-morpholin-4-ylcarbonylpiperidin-3-yl]benzimidazole-2-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.-C, Lane, W. | | Deposit date: | 2016-07-01 | | Release date: | 2017-07-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of TAK-272: A Novel, Potent and Orally Active Renin Inhibitor
To be published
|
|
1OJK
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7BE197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL, ... | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
1OLZ
 
 | | The ligand-binding face of the semaphorins revealed by the high resolution crystal structure of SEMA4D | | Descriptor: | SEMAPHORIN 4D | | Authors: | Love, C.A, Harlos, K, Mavaddat, N, Davis, S.J, Stuart, D.I, Jones, E.Y, Esnouf, R.M. | | Deposit date: | 2003-08-19 | | Release date: | 2003-09-11 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Ligand-Binding Face of the Semaphorins Revealed by the High-Resolution Crystal Structure of Sema4D
Nat.Struct.Biol., 10, 2003
|
|
1OJI
 
 | | Anatomy of glycosynthesis: Structure and kinetics of the Humicola insolens Cel7B E197A and E197S glycosynthase mutants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I, GLYCEROL | | Authors: | Ducros, V.M.-A, Tarling, C.A, Zechel, D.L, Brzozowski, A.M, Frandsen, T.P, Von Ossowski, I, Schulein, M, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-07-10 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Anatomy of Glycosynthesis: Structure and Kinetics of the Humicola Insolens Cel7B E197A and E197S Glycosynthase Mutants
Chem.Biol., 10, 2003
|
|
3PI1
 
 | | Crystallographic Structure of HbII-oxy from Lucina pectinata at pH 9.0 | | Descriptor: | Hemoglobin II, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gavira, J.A, Nieves-Marrero, C.A, Ruiz-Martinez, C.R, Estremera-Andujar, R.A, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-11-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | pH-dependence crystallographic studies of the oxygen carrier hemoglobin II from Lucina pectinata
To be Published
|
|
5KVH
 
 | | Crystal structure of human apoptosis-inducing factor with W196A mutation | | Descriptor: | Apoptosis-inducing factor 1, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
5KKZ
 
 | | Rhodobacter sphaeroides bc1 with famoxadone | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, ASCORBIC ACID, Cytochrome b, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Tang, W.K, Yu, C.A. | | Deposit date: | 2016-06-23 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
5KVI
 
 | | Crystal structure of monomeric human apoptosis-inducing factor with E413A/R422A/R430A mutations | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Apoptosis-inducing factor 1, mitochondrial, ... | | Authors: | Brosey, C.A, Nix, J, Ellenberger, T, Tainer, J.A. | | Deposit date: | 2016-07-14 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Defining NADH-Driven Allostery Regulating Apoptosis-Inducing Factor.
Structure, 24, 2016
|
|
3PI4
 
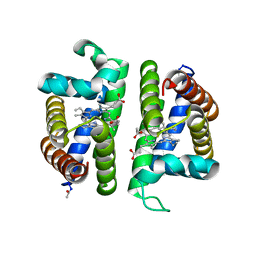 | | Crystallographic Structure of HbII-oxy from Lucina pectinata at pH 4.0 | | Descriptor: | Hemoglobin II, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gavira, J.A, Nieves-Marrero, C.A, Ruiz-Martinez, C.R, Estremera-Andujar, R.A, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | pH-dependence crystallographic studies of the oxygen carrier hemoglobin II from Lucina pectinata
To be Published
|
|
1QH1
 
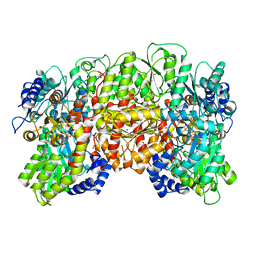 | | NITROGENASE MOFE PROTEIN FROM KLEBSIELLA PNEUMONIAE, PHENOSAFRANIN OXIDIZED STATE | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, ... | | Authors: | Mayer, S.M, Lawson, D.M, Gormal, C.A, Roe, S.M, Smith, B.E. | | Deposit date: | 1999-05-10 | | Release date: | 1999-11-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into structure-function relationships in nitrogenase: A 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein.
J.Mol.Biol., 292, 1999
|
|
1QGU
 
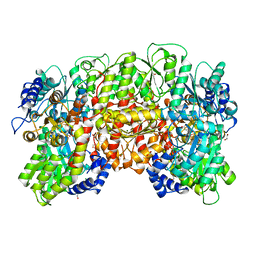 | | NITROGENASE MO-FE PROTEIN FROM KLEBSIELLA PNEUMONIAE, DITHIONITE-REDUCED STATE | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CHLORIDE ION, ... | | Authors: | Mayer, S.M, Lawson, D.M, Gormal, C.A, Roe, S.M, Smith, B.E. | | Deposit date: | 1999-05-06 | | Release date: | 1999-11-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into structure-function relationships in nitrogenase: A 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein.
J.Mol.Biol., 292, 1999
|
|
1QRJ
 
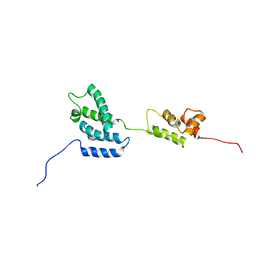 | | Solution structure of htlv-i capsid protein | | Descriptor: | HTLV-I CAPSID PROTEIN | | Authors: | Khorasanizadeh, S, Campos-Olivas, R, Clark, C.A, Summers, M.F. | | Deposit date: | 1999-06-14 | | Release date: | 1999-07-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific 1H, 13C and 15N chemical shift assignment and secondary structure of the HTLV-I capsid protein.
J.Biomol.NMR, 14, 1999
|
|
1Q0B
 
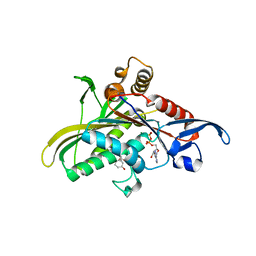 | | Crystal structure of the motor protein KSP in complex with ADP and monastrol | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ETHYL 4-(3-HYDROXYPHENYL)-6-METHYL-2-THIOXO-1,2,3,4-TETRAHYDROPYRIMIDINE-5-CARBOXYLATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y, Sardana, V, Xu, B, Halczenko, W, Homnick, C, Buser, C.A, Hartman, G.D, Huber, H.E, Kuo, L.C. | | Deposit date: | 2003-07-15 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibition of a mitotic motor protein: where, how, and conformational consequences
J.Mol.Biol., 335, 2004
|
|
1Q15
 
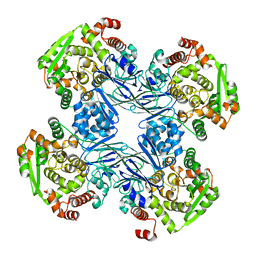 | | Carbapenam Synthetase | | Descriptor: | CarA | | Authors: | Miller, M.T, Gerratana, B, Stapon, A, Townsend, C.A, Rosenzweig, A.C. | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Carbapenam Synthetase (CarA)
J.Biol.Chem., 278, 2003
|
|
1Q4R
 
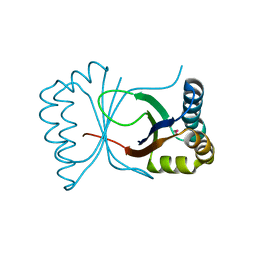 | | Gene Product of At3g17210 from Arabidopsis Thaliana | | Descriptor: | MAGNESIUM ION, protein At3g17210 | | Authors: | Phillips Jr, G.N, Bingman, C.A, Johnson, K.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-04 | | Release date: | 2003-11-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the protein from gene At3g17210 of Arabidopsis thaliana
Proteins, 57, 2004
|
|
1LAY
 
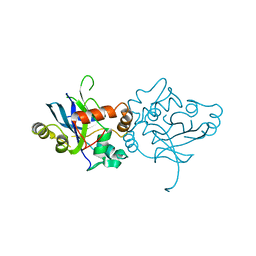 | | CRYSTAL STRUCTURE OF CYTOMEGALOVIRUS PROTEASE | | Descriptor: | CYTOMEGALOVIRUS PROTEASE | | Authors: | Qiu, X, Culp, J.S, Dilella, A.G, Hellmig, B, Hoog, S.S, Jason, C.A, Smith, W.W, Abdel-Meguid, S.S. | | Deposit date: | 1996-07-16 | | Release date: | 1997-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique fold and active site in cytomegalovirus protease.
Nature, 383, 1996
|
|
1PK8
 
 | | Crystal Structure of Rat Synapsin I C Domain Complexed to Ca.ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Brautigam, C.A, Chelliah, Y, Deisenhofer, J. | | Deposit date: | 2003-06-05 | | Release date: | 2004-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetramerization and ATP binding by a protein comprising the A, B, and C domains of rat synapsin I.
J.Biol.Chem., 279, 2004
|
|
1Q20
 
 | | Crystal Structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of PAP and pregnenolone | | Descriptor: | (3BETA)-3-HYDROXYPREGN-5-EN-20-ONE, ADENOSINE-3'-5'-DIPHOSPHATE, SODIUM ION, ... | | Authors: | Lee, K.A, Fuda, H, Lee, Y.C, Negishi, M, Strott, C.A, Pedersen, L.C. | | Deposit date: | 2003-07-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone
and 3'-phosphoadenosine 5'-phosphate. Rationale for specificity differences between prototypical
SULT2A1 and the SULT2BG1 isoforms.
J.Biol.Chem., 278, 2003
|
|
1Q45
 
 | | 12-0xo-phytodienoate reductase isoform 3 | | Descriptor: | 12-oxophytodienoate-10,11-reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Arabidopsis At2g06050, 12-oxophytodienoate reductase isoform 3
Proteins, 58, 2005
|
|
1QS8
 
 | | Crystal structure of the P. vivax aspartic proteinase plasmepsin complexed with the inhibitor pepstatin A | | Descriptor: | ACETATE ION, PEPSTATIN A, PLASMEPSIN | | Authors: | Khazanovich Bernstein, N, Cherney, M.M, Yowell, C.A, Dame, J.B, James, M.N.G. | | Deposit date: | 1999-06-25 | | Release date: | 1999-07-07 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation of P. vivax plasmepsin.
J.Mol.Biol., 329, 2003
|
|
1QE0
 
 | | CRYSTAL STRUCTURE OF APO S. AUREUS HISTIDYL-TRNA SYNTHETASE | | Descriptor: | Histidine--tRNA ligase | | Authors: | Qiu, X, Janson, C.A, Blackburn, M.N, Chohan, I.K, Hibbs, M, Abdel-Meguid, S.S. | | Deposit date: | 1999-07-12 | | Release date: | 2000-07-12 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative structural dynamics and a novel fidelity mechanism in histidyl-tRNA synthetases.
Biochemistry, 38, 1999
|
|
