1QCR
 
 | | CRYSTAL STRUCTURE OF BOVINE MITOCHONDRIAL CYTOCHROME BC1 COMPLEX, ALPHA CARBON ATOMS ONLY | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, UBIQUINOL CYTOCHROME C OXIDOREDUCTASE | | Authors: | Xia, D, Yu, C.A, Kim, H, Xia, J.Z, Kachurin, A, Zhang, L, Yu, L, Deisenhofer, J. | | Deposit date: | 1997-05-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria.
Science, 277, 1997
|
|
2BFC
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
3H5R
 
 | | Crystal structure of E. coli MccB + Succinimide | | Descriptor: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H7C
 
 | | Crystal Structure of Arabidopsis thaliana Agmatine Deiminase from Cell Free Expression | | Descriptor: | 2,2',2''-NITRILOTRIETHANOL, Agmatine deiminase, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-04-24 | | Release date: | 2009-05-26 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of Arabidopsis thaliana Agmatine Deiminase
To be Published
|
|
2BPD
 
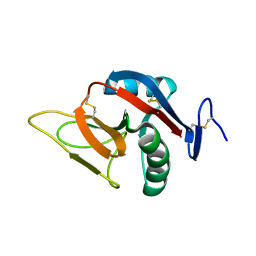 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | DECTIN-1 | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
2BPH
 
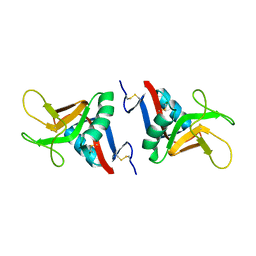 | | STRUCTURE OF MURINE DECTIN-1 | | Descriptor: | DECTIN-1, MAGNESIUM ION | | Authors: | Brown, J, O'Callaghan, C.A, Marshall, A.S.J, Gilbert, R.J.C, Siebold, C, Gordon, S, Brown, G.D, Jones, E.Y. | | Deposit date: | 2005-04-19 | | Release date: | 2006-08-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Fungal Beta-Glucan-Binding Immune Receptor Dectin-1: Implications for Function.
Protein Sci., 16, 2007
|
|
3HAV
 
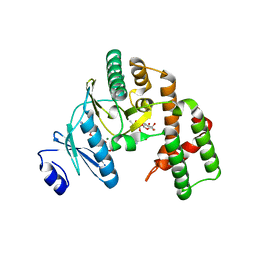 | | Structure of the streptomycin-ATP-APH(2")-IIa ternary complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aminoglycoside phosphotransferase, MAGNESIUM ION, ... | | Authors: | Young, P.G, Baker, E.N, Vakulenko, S.B, Smith, C.A. | | Deposit date: | 2009-05-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2''-phosphotransferase-IIa [APH(2'')-IIa] provide insights into substrate selectivity in the APH(2'') subfamily.
J.Bacteriol., 191, 2009
|
|
1HN9
 
 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III | | Descriptor: | BETA-KETOACYL-ACYL CARRIER PROTEIN SYNTHASE III, PHOSPHATE ION | | Authors: | Qiu, X, Janson, C.A, Konstantinidis, A.K, Nwagwu, S, Silverman, C, Smith, W.W, Khandekar, S.K, Lonsdale, J, Abdel-Meguid, S.S. | | Deposit date: | 2000-12-07 | | Release date: | 2000-12-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of beta-ketoacyl-acyl carrier protein synthase III. A key condensing enzyme in bacterial fatty acid biosynthesis.
J.Biol.Chem., 274, 1999
|
|
1R64
 
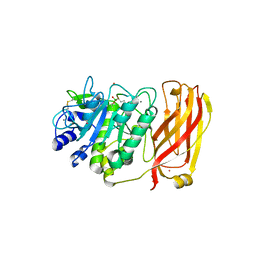 | | The 2.2 A crystal structure of Kex2 protease in complex with Ac-Arg-Glu-Lys-boroArg peptidyl boronic acid inhibitor | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Holyoak, T, Kettner, C.A, Petsko, G.A, Fuller, R.S, Ringe, D. | | Deposit date: | 2003-10-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Differences in Substrate Selectivity in Kex2 and Furin Protein Convertases
Biochemistry, 43, 2004
|
|
2BFE
 
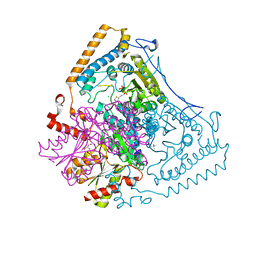 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFB
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BFD
 
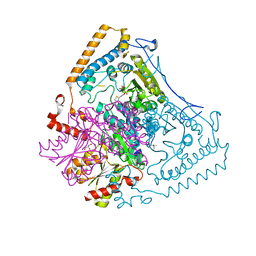 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
3H9G
 
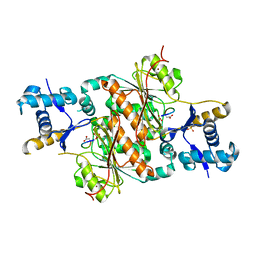 | | Crystal structure of E. coli MccB + MccA-N7isoASN | | Descriptor: | MccB protein, Microcin C7 analog, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3HAM
 
 | | Structure of the gentamicin-APH(2")-IIa complex | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, Aminoglycoside phosphotransferase, GLYCEROL | | Authors: | Young, P.G, Baker, E.N, Vakulenko, S.B, Smith, C.A. | | Deposit date: | 2009-05-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of substrate and nucleotide complexes of Enterococcus faecium aminoglycoside-2''-phosphotransferase-IIa [APH(2'')-IIa] provide insights into substrate selectivity in the APH(2'') subfamily.
J.Bacteriol., 191, 2009
|
|
3IR2
 
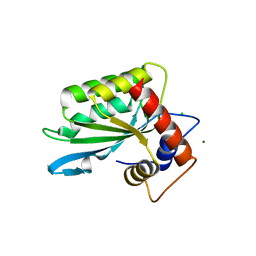 | |
2BEU
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2C6U
 
 | |
2FNS
 
 | | Crystal structure of wild-type inactive (D25N) HIV-1 protease complexed with wild-type HIV-1 NC-p1 substrate. | | Descriptor: | ACETATE ION, NC-P1 SUBSTRATE PEPTIDE, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, Schiffer, C.A. | | Deposit date: | 2006-01-11 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of substrate recognition by drug-resistant human immunodeficiency virus type 1 protease variants revealed by a novel structural intermediate.
J.Virol., 80, 2006
|
|
3IAA
 
 | | Crystal Structure of CalG2, Calicheamicin Glycosyltransferase, TDP bound form | | Descriptor: | CalG2, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Chang, A, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2009-07-13 | | Release date: | 2010-06-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Complete set of glycosyltransferase structures in the calicheamicin biosynthetic pathway reveals the origin of regiospecificity.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7SCC
 
 | | T-cylinder of Synechocystis PCC 6803 Phycobilisome, complex with OCP - local refinement | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Orange carotenoid-binding protein, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
7SCB
 
 | | B-cylinder of Synechocystis PCC 6803 Phycobilisome, complex with OCP - local refinement | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
7SCA
 
 | | Synechocystis PCC 6803 Phycobilisome rod from OCP-PBS complex sample | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
7SC9
 
 | | Synechocystis PCC 6803 Phycobilisome core, complex with OCP | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
7SC8
 
 | | Synechocystis PCC 6803 Phycobilisome rod from PBS sample | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
7SC7
 
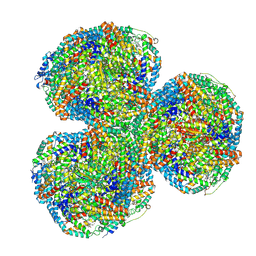 | | Synechocystis PCC 6803 Phycobilisome core from up-down rod conformation | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Dominguez-Martin, M.A, Kirst, H, Kerfeld, C.A. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of a phycobilisome in light-harvesting and photoprotected states.
Nature, 609, 2022
|
|
