4WJI
 
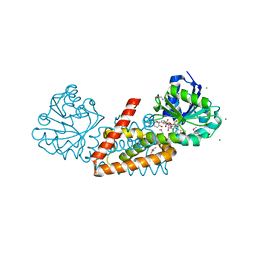 | | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP and tyrosine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Shabalin, I.G, Cooper, D.R, Hou, J, Zimmerman, M.D, Stead, M, Hillerich, B.S, Ahmed, M, Hammonds, J, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-30 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of cyclohexadienyl dehydrogenase from Sinorhizobium meliloti in complex with NADP
to be published
|
|
7NR7
 
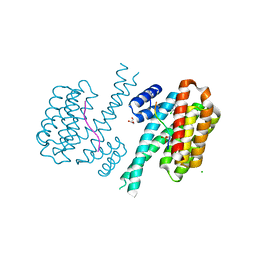 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-111 | | Descriptor: | 14-3-3 protein sigma, 4-(2-chloranyl-5-methoxy-benzimidazol-1-yl)benzaldehyde, 4-(2-chloranyl-6-methoxy-benzimidazol-1-yl)benzaldehyde, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-03-03 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O5O
 
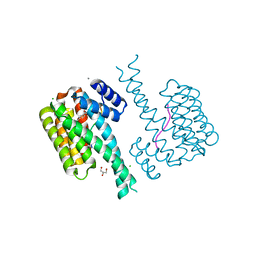 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-165 | | Descriptor: | 14-3-3 protein sigma, 4-methanoyl-~{N}-[(1-methylpyrazol-4-yl)methyl]benzamide, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-09 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7O5X
 
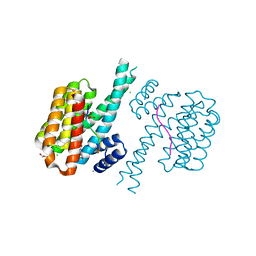 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-173 | | Descriptor: | 14-3-3 protein sigma, 4-(4-methoxypiperidin-1-yl)carbonylbenzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-09 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
8SG3
 
 | |
4WSQ
 
 | | Crystal Structure of Adaptor Protein 2 Associated Kinase (AAK1) in complex with small molecule inhibitor | | Descriptor: | 1,2-ETHANEDIOL, AP2-associated protein kinase 1, K-252A, ... | | Authors: | Sorrell, F.J, Elkins, J.M, Krojer, T, Williams, E, Abdul, K, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-10-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Family-wide Structural Analysis of Human Numb-Associated Protein Kinases.
Structure, 24, 2016
|
|
7O6I
 
 | |
7O6O
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-096 | | Descriptor: | (5-methanoyl-2-nitro-phenyl) propane-2-sulfonate, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-04-11 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NRK
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1002F1 | | Descriptor: | 14-3-3 protein sigma, 4-(4-methylimidazol-1-yl)benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-03-04 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NQP
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound LvD1009 | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-(2-phenylimidazol-1-yl)benzaldehyde, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-03-02 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
4WNJ
 
 | | Crystal structure of Transthyretin-quercetin complex | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, DIMETHYL SULFOXIDE, Transthyretin | | Authors: | Zanotti, G, Cianci, M, Berni, R, Folli, C. | | Deposit date: | 2014-10-13 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Structural evidence for asymmetric ligand binding to transthyretin.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1SOY
 
 | | Solution structure of the bacterial frataxin orthologue, CyaY | | Descriptor: | CyaY protein | | Authors: | Nair, M, Adinolfi, S, Pastore, C, Kelly, G, Temussi, P, Pastore, A. | | Deposit date: | 2004-03-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Bacterial Frataxin Ortholog, CyaY; Mapping the Iron Binding Sites
Structure, 12, 2004
|
|
4WR4
 
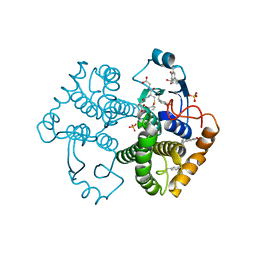 | | Crystal Structure of GST Mutated with Halogenated Tyrosine (7bGST-1) | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, SULFATE ION | | Authors: | Akasaka, R, Kawazoe, M, Tomabechi, Y, Ohtake, K, Itagaki, T, Takemoto, C, Shirouzu, M, Yokoyama, S, Sakamoto, K. | | Deposit date: | 2014-10-23 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Protein stabilization utilizing a redefined codon
Sci Rep, 5, 2015
|
|
5HYP
 
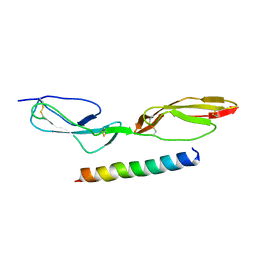 | |
4WZ9
 
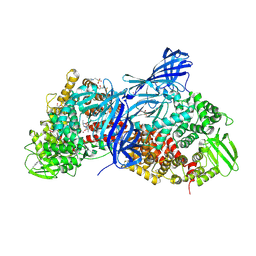 | | APN1 from Anopheles gambiae | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, AGAP004809-PA, ALA-ALA-ALA-LYS-ALA, ... | | Authors: | Atkinson, S.C, Armistead, J.S, Mathias, D.K, Sandeu, M.M, Tao, D, Borhani-Dizaji, N, Morlais, I, Dinglasan, R.R, Borg, N.A. | | Deposit date: | 2014-11-19 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Anopheles-midgut APN1 structure reveals a new malaria transmission-blocking vaccine epitope.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4WRD
 
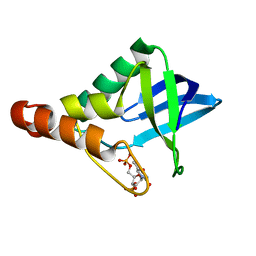 | |
1SLL
 
 | | SIALIDASE L FROM LEECH MACROBDELLA DECORA | | Descriptor: | SIALIDASE L | | Authors: | Luo, Y, Li, S.C, Chou, M.Y, Li, Y.T, Luo, M. | | Deposit date: | 1997-10-14 | | Release date: | 1998-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of an intramolecular trans-sialidase with a NeuAc alpha2-->3Gal specificity.
Structure, 6, 1998
|
|
7O4E
 
 | | The DYW domain of A. thaliana OTP86 in its inactive state | | Descriptor: | Pentatricopeptide repeat-containing protein At3g63370, chloroplastic, ZINC ION | | Authors: | Weber, G, Palm, G.J, Takenaka, M, Barthel, T, Feiler, C, Weiss, M.S. | | Deposit date: | 2021-04-06 | | Release date: | 2021-06-30 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DYW domain structures imply an unusual regulation principle in plant organellar RNA editing catalysis.
Nat Catal, 4, 2021
|
|
3MF7
 
 | | Crystal Structure of (R)-oxirane-2-carboxylate inhibited cis-CaaD | | Descriptor: | Cis-3-chloroacrylic acid dehalogenase | | Authors: | Guo, Y, Serrano, H, Ernst, S.R, Johnson Jr, W.H, Hackert, M.L, Whitman, C.P. | | Deposit date: | 2010-04-01 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of native and inactivated cis-3-chloroacrylic acid dehalogenase: Implications for the catalytic and inactivation mechanisms.
Bioorg.Chem., 39, 2011
|
|
4X4A
 
 | | Crystal structure of the intramolecular trans-sialidase from Ruminococcus gnavus in complex with 2,7-Anhydro-Neu5Ac | | Descriptor: | 2-ACETYLAMINO-7-(1,2-DIHYDROXY-ETHYL)-3-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCTANE-5-CARBOXYLIC ACID, ACETYL GROUP, Anhydrosialidase, ... | | Authors: | Owen, C.D, Tailford, L.E, Taylor, G.L, Juge, N. | | Deposit date: | 2014-12-02 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of intramolecular trans-sialidases in human gut microbiota suggests novel mechanisms of mucosal adaptation.
Nat Commun, 6, 2015
|
|
2RON
 
 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
4KMS
 
 | | Crystal structure of Acetoacetyl-CoA reductase from Rickettsia felis | | Descriptor: | Acetoacetyl-CoA reductase | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Abendroth, J, Lukacs, C, Edwards, T.E, Lorimer, D. | | Deposit date: | 2013-05-08 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of acetoacetyl-CoA reductase from Rickettsia felis.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4X8L
 
 | | Crystal structure of E. coli Adenylate kinase P177A mutant in complex with inhibitor Ap5a | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
1S8K
 
 | | Solution Structure of BmKK4, A Novel Potassium Channel Blocker from Scorpion Buthus martensii Karsch, 25 structures | | Descriptor: | Toxin BmKK4 | | Authors: | Zhang, N, Chen, X, Li, M, Cao, C, Wang, Y, Hu, G, Wu, H. | | Deposit date: | 2004-02-02 | | Release date: | 2005-02-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK4, the first member of subfamily alpha-KTx 17 of scorpion toxins
Biochemistry, 43, 2004
|
|
2RMS
 
 | | Solution structure of the mSin3A PAH1-SAP25 SID complex | | Descriptor: | MSin3A-binding protein, Paired amphipathic helix protein Sin3a | | Authors: | Sahu, S.C, Swanson, K.A, Kang, R.S, Huang, K, Brubaker, K, Ratcliff, K, Radhakrishnan, I. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conserved Themes in Target Recognition by the PAH1 and PAH2 Domains of the Sin3 Transcriptional Corepressor
J.Mol.Biol., 375, 2007
|
|
