5G3X
 
 | | Structure of recombinant granulovirus polyhedrin | | Descriptor: | GRANULOVIRUS POLYHEDRIN | | Authors: | Bunker, R.D, Chiu, E, Metcalf, P. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2FE1
 
 | | Crystal Structure of PAE0151 from Pyrobaculum aerophilum | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Bunker, R.D, Baker, E.N, Arcus, V.L. | | Deposit date: | 2005-12-15 | | Release date: | 2005-12-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PAE0151 from Pyrobaculum aerophilum, a PIN-domain (VapC) protein from a toxin-antitoxin operon.
Proteins, 72, 2008
|
|
5NW5
 
 | | Crystal structure of the Rif1 N-terminal domain (RIF1-NTD) from Saccharomyces cerevisiae in complex with DNA | | Descriptor: | DNA (30-MER), DNA (60-MER), Telomere length regulator protein RIF1 | | Authors: | Bunker, R.D, Reinert, J.K, Shi, T, Thoma, N.H. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6.502 Å) | | Cite: | Rif1 maintains telomeres and mediates DNA repair by encasing DNA ends.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5NVR
 
 | |
6YNG
 
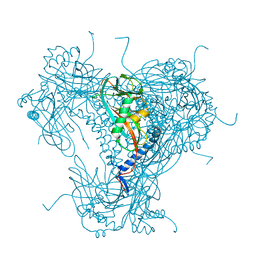 | |
6GVW
 
 | | Crystal structure of the BRCA1-A complex | | Descriptor: | BRCA1-A complex subunit Abraxas 1, BRCA1-A complex subunit RAP80, BRISC and BRCA1-A complex member 1, ... | | Authors: | Bunker, R.D, Rabl, J, Thoma, N.H. | | Deposit date: | 2018-06-21 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation.
Mol.Cell, 75, 2019
|
|
6H0G
 
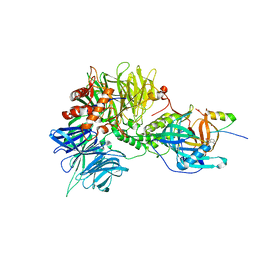 | | Structure of the DDB1-CRBN-pomalidomide complex bound to ZNF692(ZF4) | | Descriptor: | DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1,DDB1 (DNA damage binding protein 1),DNA damage-binding protein 1,DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Bunker, R.D, Petzold, G, Thoma, N.H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN.
Science, 362, 2018
|
|
6H3C
 
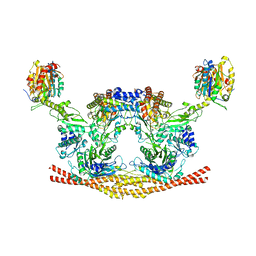 | | Cryo-EM structure of the BRISC complex bound to SHMT2 | | Descriptor: | BRISC and BRCA1-A complex member 1, BRISC and BRCA1-A complex member 2, BRISC complex subunit Abraxas 2, ... | | Authors: | Bunker, R.D, Rabl, J, Thoma, N.H. | | Deposit date: | 2018-07-18 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation.
Mol.Cell, 75, 2019
|
|
6TD3
 
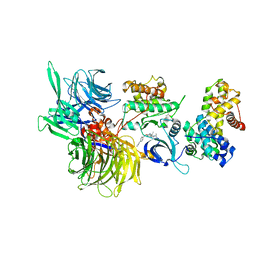 | | Structure of DDB1 bound to CR8-engaged CDK12-cyclinK | | Descriptor: | (2R)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Bunker, R.D, Petzold, G, Kozicka, Z, Thoma, N.H. | | Deposit date: | 2019-11-07 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K.
Nature, 585, 2020
|
|
4WPY
 
 | | Racemic crystal structure of Rv1738 from Mycobacterium tuberculosis (Form-II) | | Descriptor: | GLYCEROL, protein DL-Rv1738, trifluoroacetic acid | | Authors: | Bunker, R.D, Mandal, K, Kent, S.B.H, Baker, E.N. | | Deposit date: | 2014-10-21 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WSP
 
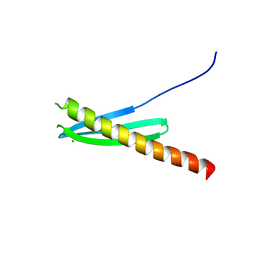 | | Racemic crystal structure of Rv1738 from Mycobacterium tuberculosis (Form-I) | | Descriptor: | CHLORIDE ION, protein DL-Rv1738 | | Authors: | Bunker, R.D, Mandal, K, Kent, S.B.H, Baker, E.N. | | Deposit date: | 2014-10-28 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A functional role of Rv1738 in Mycobacterium tuberculosis persistence suggested by racemic protein crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WSN
 
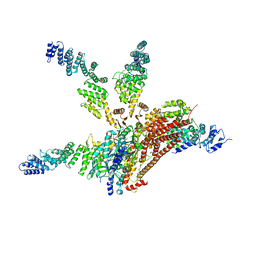 | | Crystal structure of the COP9 signalosome, a P1 crystal form | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | Deposit date: | 2014-10-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome.
Nature, 531, 2016
|
|
4BC4
 
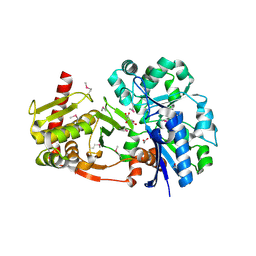 | | Crystal structure of human D-xylulokinase in complex with D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, D-XYLULOSE, XYLULOSE KINASE | | Authors: | Bunker, R.D, Loomes, K.M, Baker, E.N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Function of Human Xylulokinase, an Enzyme with Important Roles in Carbohydrate Metabolism
J.Biol.Chem., 288, 2013
|
|
4BC2
 
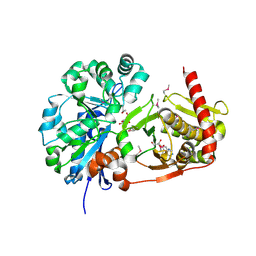 | | Crystal structure of human D-xylulokinase in complex with D-xylulose and adenosine diphosphate | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, D-XYLULOSE, ... | | Authors: | Bunker, R.D, Loomes, K.M, Baker, E.N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and Function of Human Xylulokinase, an Enzyme with Important Roles in Carbohydrate Metabolism
J.Biol.Chem., 288, 2013
|
|
4BC3
 
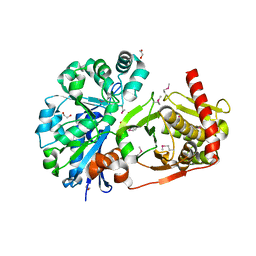 | | Crystal structure of human D-xylulokinase | | Descriptor: | 1,2-ETHANEDIOL, XYLULOSE KINASE | | Authors: | Bunker, R.D, Loomes, K.M, Baker, E.N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure and Function of Human Xylulokinase, an Enzyme with Important Roles in Carbohydrate Metabolism
J.Biol.Chem., 288, 2013
|
|
4D10
 
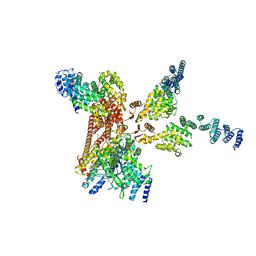 | | Crystal structure of the COP9 signalosome | | Descriptor: | COP9 SIGNALOSOME COMPLEX SUBUNIT 1, COP9 SIGNALOSOME COMPLEX SUBUNIT 2, COP9 SIGNALOSOME COMPLEX SUBUNIT 3, ... | | Authors: | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | Deposit date: | 2014-04-30 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structure of the Human Cop9 Signalosome
Nature, 512, 2014
|
|
4D18
 
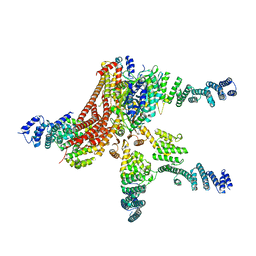 | | Crystal structure of the COP9 signalosome | | Descriptor: | COP9 SIGNALOSOME COMPLEX SUBUNIT 1, COP9 SIGNALOSOME COMPLEX SUBUNIT 2, COP9 SIGNALOSOME COMPLEX SUBUNIT 3, ... | | Authors: | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.08 Å) | | Cite: | Crystal Structure of the Human Cop9 Signalosome
Nature, 512, 2014
|
|
4D0P
 
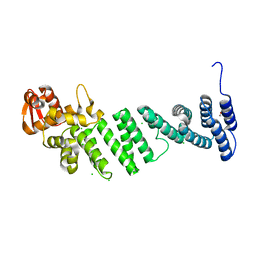 | | Crystal structure of human CSN4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COP9 SIGNALOSOME COMPLEX SUBUNIT 4, ... | | Authors: | Bunker, R.D, Lingaraju, G.M, Thoma, N.H. | | Deposit date: | 2014-04-29 | | Release date: | 2014-07-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Cop9 Signalosome
Nature, 512, 2014
|
|
4BC5
 
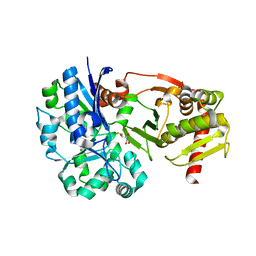 | | Crystal structure of human D-xylulokinase in complex with inhibitor 5- deoxy-5-fluoro-D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, 5-deoxy-5-fluoro-D-xylulose, XYLULOSE KINASE | | Authors: | Bunker, R.D, Loomes, K.M, Baker, E.N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Function of Human Xylulokinase, an Enzyme with Important Roles in Carbohydrate Metabolism
J.Biol.Chem., 288, 2013
|
|
4BJS
 
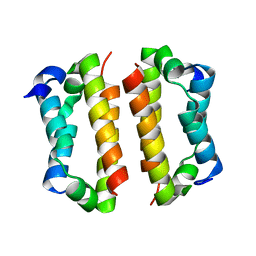 | | Crystal structure of the Rif1 C-terminal domain (Rif1-CTD) from Saccharomyces cerevisiae | | Descriptor: | TELOMERE LENGTH REGULATOR PROTEIN RIF1 | | Authors: | Bunker, R.D, Shi, T, Gut, H, Scrima, A, Thoma, N.H. | | Deposit date: | 2013-04-19 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
3KLQ
 
 | | Crystal Structure of the Minor Pilin FctB from Streptococcus pyogenes 90/306S | | Descriptor: | GLYCEROL, Putative pilus anchoring protein | | Authors: | Linke, C, Young, P.G, Bunker, R.D, Caradoc-Davies, T.T, Baker, E.N. | | Deposit date: | 2009-11-08 | | Release date: | 2010-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the minor pilin FctB reveals determinants of Group A streptococcal pilus anchoring
J.Biol.Chem., 285, 2010
|
|
5G0Z
 
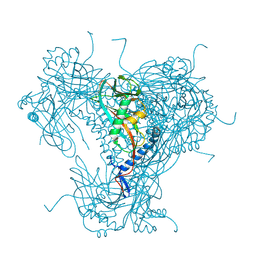 | | Structure of native granulovirus polyhedrin determined using an X-ray free-electron laser | | Descriptor: | GRANULIN | | Authors: | Gati, C, Bunker, R.D, Oberthur, D, Metcalf, P, Henry, C. | | Deposit date: | 2016-03-23 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4YN2
 
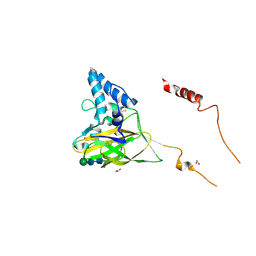 | | THE ATOMIC STRUCTURE OF WISEANA SPP ENTOMOPOXVIRUS (WSEPV) FUSOLIN SPINDLES | | Descriptor: | 1,2-ETHANEDIOL, FUSOLIN, ZINC ION, ... | | Authors: | Chiu, E, Bunker, R.D, Metcalf, P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YN1
 
 | | THE ATOMIC STRUCTURE OF ANOMALA CUPREA ENTOMOPOXVIRUS (ACEPV) FUSOLIN SPINDLES | | Descriptor: | 1,2-ETHANEDIOL, Fusolin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chiu, E, Bunker, R.D, Metcalf, P. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the enhancement of virulence by viral spindles and their in vivo crystallization.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6YOV
 
 | | OCT4-SOX2-bound nucleosome - SHL+6 | | Descriptor: | DNA (142-MER), Green fluorescent protein,POU domain, class 5, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2020-04-15 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
